Three points of consideration before testing the effect of patch connectivity on local species richness: patch delineation, scaling and variability of metrics
F. Laroche, M. Balbi, T. Grébert, F. Jabot & F. Archaux
https://doi.org/10.1101/640995
Good practice guidelines for testing species-isolation relationships in patch-matrix systems
Recommended by Damaris Zurell based on reviews by 3 anonymous reviewers
Conservation biology is strongly rooted in the theory of island biogeography (TIB). In island systems where the ocean constitutes the inhospitable matrix, TIB predicts that species richness increases with island size as extinction rates decrease with island area (the species-area relationship, SAR), and species richness increases with connectivity as colonisation rates decrease with island isolation (the species-isolation relationship, SIR)[1]. In conservation biology, patches of habitat (habitat islands) are often regarded as analogous to islands within an unsuitable matrix [2], and SAR and SIR concepts have received much attention as habitat loss and habitat fragmentation are increasingly threatening biodiversity [3,4].
The existence of SAR in patch-matrix systems has been confirmed in several studies, while the relative importance of SIR remains debated [2,5] and empirical evidence is mixed. For example, Thiele et al. [6] showed that connectivity effects are trait specific and more important to explain species richness of short-distant dispersers and of specialist species for which the matrix is less permeable. Some authors have also cautioned that the relative support for or against the existence of SIR may depend on methodological decisions related to connectivity metrics, patch classification, scaling decisions and sample size [7].
In this preprint, Laroche and colleagues [8] argue that methodological limits should be fully understood before questioning the validity of SIR in patch-matrix systems. In consequence, they used a virtual ecologist approach [9] to qualify different methodological aspects and derive good practice guidelines related to patch delineation, patch connectivity indices, and scaling of indices with species dispersal distance.
Laroche et al. [8] simulated spatially-explicit neutral meta-communities with up to 100 species in artificial fractal (patch-matrix) landscapes. Each habitat cell could hold up to 100 individuals. In each time step, some individuals died and were replaced by an individual from the regional species pool depending on relative local and regional abundance as well as dispersal distance to the nearest source habitat cell. Different scenarios were run with varying degrees of spatial autocorrelation in the fractal landscape (determining the clumpiness of habitat cells), the proportion of suitable habitat, and the species dispersal distances (with all species showing the same dispersal distance). Laroche and colleagues then sampled species richness in the simulated meta-communities, computed different local connectivity indices for the simulated landscapes (Buffer index with different radii, dIICflux index and dF index, and, finally, related species richness to connectivity.
The complex simulations allowed Laroche and colleagues [8] to test how methodological choices and landscape features may affect SIR. Overall, they found that patch delineation is crucial and should be fine enough to exclude potential within-patch dispersal limitations, and the scaling of the connectivity indices (in simplified words, the window of analyses) should be tailored to the dispersal distance of the species group. Of course, tuning the scaling parameters will be more complicated when dispersal distances vary across species but overall these results corroborate empirical findings that SIR effects are trait specific [6]. Additionally, the results by Laroche and colleagues [8] indicated that indices based on Euclidian rather than topological distance are more performant and that evidence of SIR is more likely if Buffer indices are highly variable between sampled patches.
Although the study is very technical due to the complex simulation approach and the different methods tested, I hope it will not only help guiding methodological choices but also inspire ecologists to further test or even revisit SIR (and SAR) hypotheses for different systems. Also, Laroche and colleagues propose many interesting avenues that could still be explored in this context, for example determining the optimal grid resolution for the patch delineation in empirical studies.
References
[1] MacArthur, R.H. and Wilson, E.O. (1967) The theory of island biogeography. Princeton University Press, Princeton.
[2] Fahrig, L. (2013) Rethinking patch size and isolation effects: the habitat amount hypothesis. Journal of Biogeography, 40(9), 1649-1663. doi: 10.1111/jbi.12130
[3] Hanski, I., Zurita, G.A., Bellocq, M.I. and Rybicki J (2013) Species–fragmented area relationship. Proceedings of the National Academy of Sciences U.S.A., 110(31), 12715-12720. doi: 10.1073/pnas.1311491110
[4] Giladi, I., May, F., Ristow, M., Jeltsch, F. and Ziv, Y. (2014) Scale‐dependent species–area and species–isolation relationships: a review and a test study from a fragmented semi‐arid agro‐ecosystem. Journal of Biogeography, 41(6), 1055-1069. doi: 10.1111/jbi.12299
[5] Hodgson, J.A., Moilanen, A., Wintle, B.A. and Thomas, C.D. (2011) Habitat area, quality and connectivity: striking the balance for efficient conservation. Journal of Applied Ecology, 48(1), 148-152. doi: 10.1111/j.1365-2664.2010.01919.x
[6] Thiele, J., Kellner, S., Buchholz, S., and Schirmel, J. (2018) Connectivity or area: what drives plant species richness in habitat corridors? Landscape Ecology, 33, 173-181. doi: 10.1007/s10980-017-0606-8
[7] Vieira, M.V., Almeida-Gomes, M., Delciellos, A.C., Cerqueira, R. and Crouzeilles, R. (2018) Fair tests of the habitat amount hypothesis require appropriate metrics of patch isolation: An example with small mammals in the Brazilian Atlantic Forest. Biological Conservation, 226, 264-270. doi: 10.1016/j.biocon.2018.08.008
[8] Laroche, F., Balbi, M., Grébert, T., Jabot, F. and Archaux, F. (2020) Three points of consideration before testing the effect of patch connectivity on local species richness: patch delineation, scaling and variability of metrics. bioRxiv, 640995, ver. 5 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/640995
[9] Zurell, D., Berger, U., Cabral, J.S., Jeltsch, F., Meynard, C.N., Münkemüller, T., Nehrbass, N., Pagel, J., Reineking, B., Schröder, B. and Grimm, V. (2010) The virtual ecologist approach: simulating data and observers. Oikos, 119(4), 622-635. doi: 10.1111/j.1600-0706.2009.18284.x
| Three points of consideration before testing the effect of patch connectivity on local species richness: patch delineation, scaling and variability of metrics | F. Laroche, M. Balbi, T. Grébert, F. Jabot & F. Archaux | <p>The Theory of Island Biogeography (TIB) promoted the idea that species richness within sites depends on site connectivity, i.e. its connection with surrounding potential sources of immigrants. TIB has been extended to a wide array of fragmented... | 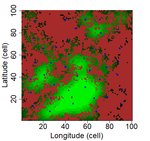 | Biodiversity, Community ecology, Dispersal & Migration, Landscape ecology, Spatial ecology, Metacommunities & Metapopulations | Damaris Zurell | | 2019-05-20 16:03:47 | View |
Comparing statistical and mechanistic models to identify the drivers of mortality within a rear-edge beech population
Cathleen Petit-Cailleux, Hendrik Davi, François Lefevre, Christophe Hurson, Joseph Garrigue, Jean-André Magdalou, Elodie Magnanou and Sylvie Oddou-Muratorio
https://doi.org/10.1101/645747
The complexity of predicting mortality in trees
Recommended by Lucía DeSoto based on reviews by Lisa Hülsmann and 2 anonymous reviewers
One of the main issues of forest ecosystems is rising tree mortality as a result of extreme weather events (Franklin et al., 1987). Eventually, tree mortality reduces forest biomass (Allen et al., 2010), although its effect on forest ecosystem fluxes seems not lasting too long (Anderegg et al., 2016). This controversy about the negative consequences of tree mortality is joined to the debate about the drivers triggering and the mechanisms accelerating tree decline. For instance, there is still room for discussion about carbon starvation or hydraulic failure determining the decay processes (Sevanto et al., 2014) or about the importance of mortality sources (Reichstein et al., 2013). Therefore, understanding and predicting tree mortality has become one of the challenges for forest ecologists in the last decade, doubling the rate of articles published on the topic (*). Although predicting the responses of ecosystems to environmental change based on the traits of species may seem a simplistic conception of ecosystem functioning (Sutherland et al., 2013), identifying those traits that are involved in the proneness of a tree to die would help to predict how forests will respond to climate threatens.
Modelling tree mortality is complex, involving multiple factors acting simultaneously at different scales, from tree genetics to ecosystem dynamics and from microsite conditions to global climatic events. Therefore, taking into account different approaches to reduce uncertainty of the predictions is needed (Bugmann et al., 2019). Petit-Cailleux et al. (2020) uses statistical and process-based models to detect the main mortality drivers of a drought- and frost-prone beech population. Particularly, they assessed the intra-individual characteristics of the population, that may play a decisive role explaining the differences in tree vulnerability to extreme weather events. Comparing the results of both analytical approaches, they find out several key factors, such as defoliation, leaf phenology and tree size, that were consistent between them. Even more, the process-based model showed the physiological mechanisms that may explain the individual vulnerability, for instance higher loss of hydraulic conductance may increase the mortality risk of trees with early budburst phenology and large stem diameter. The authors also successfully model annual mortality rate with a linear relationship including only three parameters: loss of conductance, biomass of reserves and late frost days.
This valuable study is a good example of the complexity in understanding and predicting tree mortality. The authors carried out the ambitious commitment of studying the inter-annual variation in mortality with 14-year dataset. However, it might be not enough time to control for the dependence of temporal data to soundly model mortality rate. The authors also acknowledge that the use of two approaches increases the knowledge from different perspectives, but at the same time comparing their results is difficult because the parameters used are not identical. Particularly, process-based models tend to consider the same microclimatic conditions for every tree in the population, and may produce inconsistences with statistical models. Alternatively, individual-based modelling might overcome some of the incompatibilities between the approaches (Zhu et al., 2019).
(*) Number (and percentage) of articles found in Web of Sciences after searching (December the 10th, 2020) “tree mortality”: from 163 (0.006%) in 2010 to 412 (0.013%) in 2020.
References
Allen et al. (2010). A global overview of drought and heat-induced tree mortality reveals emerging climate change risks for forests. Forest ecology and management, 259(4), 660-684. doi: https://doi.org/10.1016/j.foreco.2009.09.001
Anderegg et al. (2016). When a tree dies in the forest: scaling climate-driven tree mortality to ecosystem water and carbon fluxes. Ecosystems, 19(6), 1133-1147. doi: https://doi.org/10.1007/s10021-016-9982-1
Bugmann et al. (2019). Tree mortality submodels drive simulated long‐term forest dynamics: assessing 15 models from the stand to global scale. Ecosphere, 10(2), e02616. doi: https://doi.org/10.1002/ecs2.2616
Franklin, J. F., Shugart, H. H. and Harmon, M. E. (1987) Death as an ecological process: the causes, consequences, and variability of tree mortality. BioScience, 37, 550–556. doi: https://doi.org/10.2307/1310665
Petit-Cailleux, C., Davi, H., Lefèvre, F., Garrigue, J., Magdalou, J.-A., Hurson, C., Magnanou, E. and Oddou-Muratorio, S. (2020) Comparing statistical and mechanistic models to identify the drivers of mortality within a rear-edge beech population. bioRxiv, 645747, ver 7 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/645747
Reichstein et al. (2013). Climate extremes and the carbon cycle. Nature, 500(7462), 287-295. doi: https://doi.org/10.1038/nature12350
Sevanto, S., Mcdowell, N. G., Dickman, L. T., Pangle, R., and Pockman, W. T. (2014). How do trees die? A test of the hydraulic failure and carbon starvation hypotheses. Plant, cell & environment, 37(1), 153-161. doi: https://doi.org/10.1111/pce.12141
Sutherland et al. (2013). Identification of 100 fundamental ecological questions. Journal of ecology, 101(1), 58-67. doi: https://doi.org/10.1111/1365-2745.12025
Zhu, Y., Liu, Z., and Jin, G. (2019). Evaluating individual-based tree mortality modeling with temporal observation data collected from a large forest plot. Forest Ecology and Management, 450, 117496. doi: https://doi.org/10.1016/j.foreco.2019.117496
| Comparing statistical and mechanistic models to identify the drivers of mortality within a rear-edge beech population | Cathleen Petit-Cailleux, Hendrik Davi, François Lefevre, Christophe Hurson, Joseph Garrigue, Jean-André Magdalou, Elodie Magnanou and Sylvie Oddou-Muratorio | <p>Since several studies have been reporting an increase in the decline of forests, a major issue in ecology is to better understand and predict tree mortality. The interactions between the different factors and the physiological processes giving ... |  | Climate change, Physiology, Population ecology | Lucía DeSoto | | 2019-05-24 11:37:38 | View |
Deer slow down litter decomposition by reducing litter quality in a temperate forest
Simon Chollet, Morgane Maillard, Juliane Schorghuber, Sue Grayston, Jean-Louis Martin
https://doi.org/10.1101/690032
Disentangling effects of large herbivores on litter decomposition
Recommended by Sébastien Barot based on reviews by 2 anonymous reviewers
Aboveground – belowground interactions is a fascinating field that has developed in ecology since about 20 years [1]. This field has been very fruitful as measured by the numerous articles published but also by the particular role it has played in the development of soil ecology. While soil ecology has for a long time developed partially independently from “general ecology” [2], the field of aboveground – belowground interactions has shown that all ecological interactions occurring within the soil are likely to impact plant growth and plant physiology because they have their roots within the soil. In turns, this should impact the aerial system of plants (higher or lower biomasses, changes in leaf quality…), which should cascade on the aboveground food web. Conversely, all ecological interactions occurring aboveground likely impact plant growth, which should cascade to their root systems, and thus to the soil functioning and the soil food web (through changes in the emission of exudates or inputs of dead roots…). Basically, plants are linking the belowground and aboveground worlds because, as terrestrial primary producers, they need to have (1) leaves to capture CO2 and exploit light and (2) roots to absorb water and mineral nutrients. The article I presently recommend [3] tackles this general issue through the prism of the impact of large herbivores on the decomposition of leaf litter.
This issue is a relatively old one [4, 5] but still deserves efforts because there have been relatively few studies on the subject and because the issue is relatively complex due to the diversity of mechanisms involved and the difficulty to disentangle them. I recommend this article because the authors have cleverly taken advantage of a ‘‘natural’’ long-term experiment, i.e. three islands with contrasted deer densities, to test whether these large mammals are able to impact leaf litter decomposition and whether they are able to do so through changes in litter quality (because they browse the vegetation) or through changes in soil characteristics (either physical or chemical characteristics or the composition of the decomposer community). They have found that deer decrease litter decomposition, mainly through a decrease in litter quality (increase in its C:N ratio). I particularly appreciate the combination of statistics achieved to test the different hypotheses and the fair and in-depth discussion of the results.
I have to confess that I have two small regrets with this work. First, all replications are implemented within the same three islands, so that it cannot be fully excluded that measured effects should not be attributed to any other possible difference between the three islands. I am fairly sure this is not the case (at least because the three islands have the same environments) but I hope that future studies or meta-analyses will be able analyse independent deer density treatments. Second, as a soil ecologist, I am eager to see results on the decomposer communities, both microorganisms and macrofauna, of the three islands.
References
[1] Hooper, D. U., Bignell, D. E., Brown, V. K., Brussard, L., Dangerfield, J. M., Wall, D. H. and Wolters, V. (2000). Interactions between Aboveground and Belowground Biodiversity in Terrestrial Ecosystems: Patterns, Mechanisms, and Feedbacks. BioScience, 50(12), 1049-1061. doi: 10.1641/0006-3568(2000)050[1049:ibaabb]2.0.co;2
[2] Barot, S., Blouin, M., Fontaine, S., Jouquet, P., Lata, J.-C., and Mathieu, J. (2007). A Tale of Four Stories: Soil Ecology, Theory, Evolution and the Publication System. PLOS ONE, 2(11), e1248. doi: 10.1371/journal.pone.0001248
[3] Chollet S., Maillard M., Schörghuber J., Grayston S. and Martin J.-L. (2019). Deer slow down litter decomposition by reducing litter quality in a temperate forest. bioRxiv, 690032, ver. 3 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/690032
[4] Wardle, D. A., Barker, G. M., Yeates, G. W., Bonner, K. I., and Ghani, A. (2001). Introduced browsing mammals in New Zealand natural forests: aboveground and belowground consequences. Ecological Monographs, 71(4), 587-614. doi: 10.1890/0012-9615(2001)071[0587:ibminz]2.0.co;2
[5] Bardgett, R. D., and Wardle, D. A. (2003). Herbivore-mediated linkages between aboveground and belowground communities. Ecology, 84(9), 2258-2268. doi: 10.1890/02-0274
| Deer slow down litter decomposition by reducing litter quality in a temperate forest | Simon Chollet, Morgane Maillard, Juliane Schorghuber, Sue Grayston, Jean-Louis Martin | <p>In temperate forest ecosystems, the role of deer in litter decomposition, a key nutrient cycling process, remains debated. Deer may modify the decomposition process by affecting plant cover and thus modifying litter abundance. They can also alt... |  | Community ecology, Ecosystem functioning, Herbivory, Soil ecology | Sébastien Barot | | 2019-07-04 14:30:19 | View |
How would variation in environmental predictability affect the use of different learning mechanisms in a social bird?
Recommended by Aliza le Roux based on reviews by Matthew Petelle and 1 anonymous reviewer
In their pre-registered paper [1], McCune and colleagues propose a field-based study of social versus individual learning mechanisms in an avian species (great-tailed grackles) that has been expanding its geographic range. The study forms part of a longer-term project that addresses various aspects of this species’ behaviour and biology, and the experience of the team is clear from the preprint. Assessing variation in learning mechanisms in different sections of the grackles’ distribution range, the researchers will investigate how individual learning and social transmission may impact learning about novel challenges in the environment. Considering that this is a social species, the authors expect both individual learning and social transmission to occur, when groups of grackles encounter new challenges/ opportunities in the wild. This in itself is not a very unusual idea to test [2, 3], but the authors are rigorously distinguishing between imitation, emulation, local enhancement, and social enhancement. Such rigour is certainly valuable in studies of cognition in the wild.
Further, the authors predict that the contribution of individual versus social learning could vary between populations, as the core may contain fewer unfamiliar/novel stimuli than the edge, where artificial sources of water (for example) may be more common. They make an argument that the core, middle, and edge populations would experience differing levels of environmental predictability. If true, their field experiments could yield very novel results on how changes in environmental predictability affect social/individual learning in a single study species. Their data would then give unusual insights into the ecological value of individual learning and distinct forms of social learning – something that is not easy to test in wild animals. The authors consider a variety of alternative hypotheses that may ultimately explain their findings, and clarify their methods and analyses in fine detail. The authors also set out limitations clearly, and give a thorough account of their approaches and thinking.
The reviewers and I have a still-unanswered question, which is central to the study: what is the predictability or unpredictability of the core versus edge environments? Although the authors have explained similarities and distinctions between the different sections of the grackles’ range, their description feels a bit vague -- it's not as rigorous or well-defined as the rest of the paper. Such a lack of definition may be inevitable in the limitations of a preprint, but ultimately it does suggest that there may be real uncertainty about the qualitative differences between the core, edge, and middle environments. The authors do explain that a lack of variation in individual responses to the field experiments would preclude the testing of further hypothesis, but do not mention how a salient lack of variation in novelty/ predictability between the environments could impact their hypotheses.
An assessment/quantification of the rate at which the different populations of grackles encounter novel stimuli would be a cornerstone of the success of this proposed study. Certainly, the authors cannot address this in much more detail during the preprint stage, but they need to consider how to best assess/describe differences before starting the full study. Such an assessment could take the form of either a GIS desktop study (comparing, for example, rates of dam/canal construction in core versus edge sections of the distribution range), or observational/ movement data contrasting how frequently members of core versus edge populations encounter artificial sources of water/food in a given month/year. Considering the long-term nature of the larger project, it is possible that these data are already available, but I am speculating. I would highly recommend that such an assessment be undertaken, beyond the mere mention of expected differences. This would solidify the central idea that there are concrete differences between the environments.
Despite this concern, the authors attended well to the comments and recommendations of the two reviewers – both experts in cognitive ecology. It is a preprint showing clear thinking and a consideration of most of the challenges that may be encountered during the course of the study. My own opinion and the estimations of the two reviewers all underscore the originality and value of this project – this should be a very valuable and potentially novel study. I look forward to seeing the outcomes of the research.
References
[1] McCune, K. B., McElreath, R., and Logan, C. J. (2019). Investigating the use of learning mechanisms in a species that is rapidly expanding its geographic range. In principle recommendation by Peer Community In Ecology. corinalogan.com/Preregistrations/g_sociallearning.html
[2] Benson-Amram, S. and Holekamp, K. E. (2012). Innovative problem solving by wild spotted hyenas. Proceedings of the Royal Society B: Biological Sciences, 279(1744), 4087–4095. doi: 10.1098/rspb.2012.1450
[3] Federspiel, I. G., Boeckle, M., von Bayern, A. M. P. and Emery, N. J. (2019). Exploring individual and social learning in jackdaws (Corvus monedula). Learning & Behavior, 47(3), 258–270. doi: 10.3758/s13420-019-00383-8
| Investigating the use of learning mechanisms in a species that is rapidly expanding its geographic range | Kelsey McCune, Richard McElreath, Corina Logan | This is one of many studies planned for our long-term research on the role of behavior and learning in rapid geographic range expansions. Project background: Behavioral flexibility, the ability to change behavior when circumstances change based on... |  | Behaviour & Ethology, Eco-evolutionary dynamics, Foraging, Preregistrations, Social structure, Spatial ecology, Metacommunities & Metapopulations, Zoology | Aliza le Roux | | 2019-07-23 18:45:20 | View |
Investigating sex differences in genetic relatedness in great-tailed grackles in Tempe, Arizona to infer potential sex biases in dispersal
August Sevchik, Corina Logan, Melissa Folsom, Luisa Bergeron, Aaron Blackwell, Carolyn Rowney, Dieter Lukas
http://corinalogan.com/Preregistrations/gdispersal.html
Investigate fine scale sex dispersal with spatial and genetic analyses
Recommended by Sophie Beltran-Bech based on reviews by Sylvine Durand and 1 anonymous reviewer based on reviews by Sylvine Durand and 1 anonymous reviewer
The preregistration "Investigating sex differences in genetic relatedness in great-tailed grackles in Tempe, Arizona to infer potential sex biases in dispersal" [1] presents the analysis plan that will be used to genetically and spatially investigate sex-biased dispersal in great-tailed grackles (Quiscalus mexicanus).
Several hypotheses implying mating systems, intrasexual competition or sex-related handicaps have been proposed to explain the diversity of dispersal patterns between or within species according to their ecological requirements, environmental factors such as seasonality [2], or individual characteristics such as age [3] or sex [4].
In birds, females are classically the dispersing sex, while males remain close to the place they were hatched [5], with potential benefits that males derive from knowing the local environment to establish territories [6].
In great-tailed grackles the males hold territories and the females choose which territory to place their nest in [7]. In this context, the main hypothesis is that females are the dispersing sex in this species. The authors of this preregistration plan to investigate this hypothesis and its 3 alternatives ((i) the males are the dispersing sex, (ii) both sexes disperse or (iii) neither of the two sexes disperse), investigating the spatial distribution of genetic relatives.
The authors plan to measure the genetic relatedness (using SNP markers) and geographic distances among all female dyads and among all male dyads in the fine geographic scale (Tempe campus, Arizona). If females disperse away from relatives, the females will be less likely to be found geographically close to genetic relatives.
This pre-registration shows that the authors are well aware of the possible limitations of their study, particularly in relation to their population of 57 individuals, on a small scale. But they will use methods that should be able to detect a signal. They were very good at incorporating the reviewers' comments and suggestions, which enabled them to produce a satisfactory and interesting version of the manuscript presenting their hypotheses, limitations and the methods they plan to use. Another point I would like to stress is that this pre-registration practice is a very good one that makes it possible to anticipate the challenges and the type of analyses to be carried out, in particular by setting out the working hypotheses and confronting them (as well as the methods envisaged) with peers from this stage. I therefore recommend this manuscript and thank all the contributors (authors and reviewers) for their work. I look forward to seeing the outcomes of this study.
References
[1] Sevchik A., Logan C. J., Folsom M., Bergeron L., Blackwell A., Rowney C., and Lukas D. (2019). Investigating sex differences in genetic relatedness in great-tailed grackles in Tempe, Arizona to infer potential sex biases in dispersal. In principle recommendation by Peer Community In Ecology. corinalogan.com/Preregistrations/gdispersal.html
[2] Fies, M. L., Puckett, K. M., and Larson-Brogdon, B. (2002). Breeding season movements and dispersal of Northern Bobwhites in fragmented habitats of Virginia. Vol. 5 , Article 35. Available at: trace.tennessee.edu/nqsp/vol5/iss1/35
[3] Marvá, M., and San Segundo, F. (2018). Age-structure density-dependent fertility and individuals dispersal in a population model. Mathematical biosciences, 300, 157-167. doi: 10.1016/j.mbs.2018.03.029
[4] Trochet, A., Courtois, E. A., Stevens, V. M., Baguette, M., Chaine, A., Schmeller, D. S., Clobert, J., and Wiens, J. J. (2016). Evolution of sex-biased dispersal. The Quarterly Review of Biology, 91(3), 297-320. doi: 10.1086/688097
[5] Greenwood, P. J., and Harvey, P. H. (1982). The natal and breeding dispersal of birds. Annual review of ecology and systematics, 13(1), 1-21. doi: 10.1146/annurev.es.13.110182.000245
[6] Greenwood, P. J. (1980). Mating systems, philopatry and dispersal in birds and mammals. Animal behaviour, 28(4), 1140-1162. doi: 10.1016/S0003-3472(80)80103-5
[7] Johnson, K., DuVal, E., Kielt, M., and Hughes, C. (2000). Male mating strategies and the mating system of great-tailed grackles. Behavioral Ecology, 11(2), 132-141. doi: 10.1093/beheco/11.2.132
| Investigating sex differences in genetic relatedness in great-tailed grackles in Tempe, Arizona to infer potential sex biases in dispersal | August Sevchik, Corina Logan, Melissa Folsom, Luisa Bergeron, Aaron Blackwell, Carolyn Rowney, Dieter Lukas | In most bird species, females disperse prior to their first breeding attempt, while males remain close to the place they were hatched for their entire lives (Greenwood and Harvey (1982)). Explanations for such female bias in natal dispersal have f... |  | Behaviour & Ethology, Life history, Preregistrations, Social structure, Zoology | Sophie Beltran-Bech | | 2019-07-24 12:47:07 | View |
Intraspecific difference among herbivore lineages and their host-plant specialization drive the strength of trophic cascades
Arnaud Sentis, Raphaël Bertram, Nathalie Dardenne, Jean-Christophe Simon, Alexandra Magro, Benoit Pujol, Etienne Danchin and Jean-Louis Hemptinne
https://doi.org/10.1101/722140
Tell me what you’ve eaten, I’ll tell you how much you’ll eat (and be eaten)
Recommended by Sara Magalhães and Raul Costa-Pereira based on reviews by Bastien Castagneyrol and 1 anonymous reviewer
Tritrophic interactions have a central role in ecological theory and applications [1-3]. Particularly, systems comprised of plants, herbivores and predators have historically received wide attention given their ubiquity and economic importance [4]. Although ecologists have long aimed to understand the forces that govern alternating ecological effects at successive trophic levels [5], several key open questions remain (at least partially) unanswered [6]. In particular, the analysis of complex food webs has questioned whether ecosystems can be viewed as a series of trophic chains [7,8]. Moreover, whether systems are mostly controlled by top-down (trophic cascades) or bottom-up processes remains an open question [6].
Traditionally, studies have addressed how species diversity at different food chain compartments affect the strength and direction of trophic cascades [9]. For example, many studies have tested whether biological control was more efficient with more than one species of natural enemies [10-12]. Much less attention has been given to the role of within-species variation in shaping trophic cascades [13]. In particular, whereas the impact of trait variation within species of plants or predators on successive trophic levels has been recently addressed [14,15], the impact of intraspecific herbivore variation is in its infancy (but see [16]). This is at odds with the resurgent acknowledgment of the importance of individual variation for several ecological processes operating at higher levels of biological organization [17].
Sources of variation within species can come in many flavours. In herbivores, striking ecological variation can be found among populations occurring on different host plants, which become genetically differentiated, thus forming host races [18,19]. Curiously, the impact of variation across host races on the strength of trophic cascades has, to date, not been explored. This is the gap that the manuscript by Sentis and colleagues [20] fills. They experimentally studied a curious tri-trophic system where the primary consumer, pea aphids, specializes in different plant hosts, creating intraspecific variation across biotypes. Interestingly, there is also ecological variation across lineages from the same biotype. The authors set up experimental food chains, where pea aphids from different lineages and biotypes were placed in their universal legume host (broad bean plants) and then exposed to a voracious but charming predator, ladybugs. The full factorial design of this experiment allowed the authors to measure vertical effects of intraspecific variation in herbivores on both plant productivity (top-down) and predator individual growth (bottom-up).
The results nicely uncover the mechanisms by which intraspecific differences in herbivores precipitates vertical modulation in food chains. Herbivore lineage and host-plant specialization shaped the strength of trophic cascades, but curiously these effects were not modulated by density-dependence. Further, ladybugs consuming pea aphids from different lineages and biotypes grew at distinct rates, revealing bottom-up effects of intraspecific variation in herbivores.
These findings are novel and exciting for several reasons. First, they show how intraspecific variation in intermediate food chain compartments can simultaneously reverberate both top-down and bottom-up effects. Second, they bring an evolutionary facet to the understanding of trophic cascades, providing valuable insights on how genetically differentiated populations play particular ecological roles in food webs. Finally, Sentis and colleagues’ findings [20] have critical implications well beyond their study systems. From an applied perspective, they provide an evident instance on how consumers’ evolutionary specialization matters for their role in ecosystems processes (e.g. plant biomass production, predator conversion rate), which has key consequences for biological control initiatives and invasive species management. From a conceptual standpoint, their results ignite the still neglected value of intraspecific variation (driven by evolution) in modulating the functioning of food webs, which is a promising avenue for future theoretical and empirical studies.
References
[1] Price, P. W., Bouton, C. E., Gross, P., McPheron, B. A., Thompson, J. N., & Weis, A. E. (1980). Interactions among three trophic levels: influence of plants on interactions between insect herbivores and natural enemies. Annual review of Ecology and Systematics, 11(1), 41-65. doi: 10.1146/annurev.es.11.110180.000353
[2] Olff, H., Brown, V.K. & Drent, R.H. (1999). Herbivores: between plants and predators. Blackwell Science, Oxford.
[3] Tscharntke, T. & Hawkins, B.A. (2002). Multitrophic level interactions. Cambridge University Press. doi: 10.1017/CBO9780511542190
[4] Agrawal, A. A. (2000). Mechanisms, ecological consequences and agricultural implications of tri-trophic interactions. Current opinion in plant biology, 3(4), 329-335. doi: 10.1016/S1369-5266(00)00089-3
[5] Pace, M. L., Cole, J. J., Carpenter, S. R., & Kitchell, J. F. (1999). Trophic cascades revealed in diverse ecosystems. Trends in ecology & evolution, 14(12), 483-488. doi: 10.1016/S0169-5347(99)01723-1
[6] Abdala‐Roberts, L., Puentes, A., Finke, D. L., Marquis, R. J., Montserrat, M., Poelman, E. H., ... & Mooney, K. (2019). Tri‐trophic interactions: bridging species, communities and ecosystems. Ecology letters, 22(12), 2151-2167. doi: 10.1111/ele.13392
[7] Polis, G.A. & Winemiller, K.O. (1996). Food webs. Integration of patterns and dynamics. Chapmann & Hall, New York. doi: 10.1007/978-1-4615-7007-3
[8] Torres‐Campos, I., Magalhães, S., Moya‐Laraño, J., & Montserrat, M. (2020). The return of the trophic chain: Fundamental vs. realized interactions in a simple arthropod food web. Functional Ecology, 34(2), 521-533. doi: 10.1111/1365-2435.13470
[9] Polis, G. A., Sears, A. L., Huxel, G. R., Strong, D. R., & Maron, J. (2000). When is a trophic cascade a trophic cascade?. Trends in Ecology & Evolution, 15(11), 473-475. doi: 10.1016/S0169-5347(00)01971-6
[10] Sih, A., Englund, G., & Wooster, D. (1998). Emergent impacts of multiple predators on prey. Trends in ecology & evolution, 13(9), 350-355. doi: 10.1016/S0169-5347(98)01437-2
[11] Diehl, E., Sereda, E., Wolters, V., & Birkhofer, K. (2013). Effects of predator specialization, host plant and climate on biological control of aphids by natural enemies: a meta‐analysis. Journal of Applied Ecology, 50(1), 262-270. doi: 10.1111/1365-2664.12032
[12] Snyder, W. E. (2019). Give predators a complement: conserving natural enemy biodiversity to improve biocontrol. Biological control, 135, 73-82. doi: 10.1016/j.biocontrol.2019.04.017
[13] Des Roches, S., Post, D. M., Turley, N. E., Bailey, J. K., Hendry, A. P., Kinnison, M. T., ... & Palkovacs, E. P. (2018). The ecological importance of intraspecific variation. Nature Ecology & Evolution, 2(1), 57-64. doi: 10.1038/s41559-017-0402-5
[14] Bustos‐Segura, C., Poelman, E. H., Reichelt, M., Gershenzon, J., & Gols, R. (2017). Intraspecific chemical diversity among neighbouring plants correlates positively with plant size and herbivore load but negatively with herbivore damage. Ecology Letters, 20(1), 87-97. doi: 10.1111/ele.12713
[15] Start, D., & Gilbert, B. (2017). Predator personality structures prey communities and trophic cascades. Ecology letters, 20(3), 366-374. doi: 10.1111/ele.12735
[16] Turcotte, M. M., Reznick, D. N., & Daniel Hare, J. (2013). Experimental test of an eco-evolutionary dynamic feedback loop between evolution and population density in the green peach aphid. The American Naturalist, 181(S1), S46-S57. doi: 10.1086/668078
[17] Bolnick, D. I., Amarasekare, P., Araújo, M. S., Bürger, R., Levine, J. M., Novak, M., ... & Vasseur, D. A. (2011). Why intraspecific trait variation matters in community ecology. Trends in ecology & evolution, 26(4), 183-192. doi: 10.1016/j.tree.2011.01.009
[18] Drès, M., & Mallet, J. (2002). Host races in plant–feeding insects and their importance in sympatric speciation. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 357(1420), 471-492. doi: 10.1098/rstb.2002.1059
[19] Magalhães, S., Forbes, M. R., Skoracka, A., Osakabe, M., Chevillon, C., & McCoy, K. D. (2007). Host race formation in the Acari. Experimental and Applied Acarology, 42(4), 225-238. doi: 10.1007/s10493-007-9091-0
[20] Sentis, A., Bertram, R., Dardenne, N., Simon, J.-C., Magro, A., Pujol, B., Danchin, E. and J.-L. Hemptinne (2020) Intraspecific difference among herbivore lineages and their host-plant specialization drive the strength of trophic cascades. bioRxiv, 722140, ver. 4 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/722140
| Intraspecific difference among herbivore lineages and their host-plant specialization drive the strength of trophic cascades | Arnaud Sentis, Raphaël Bertram, Nathalie Dardenne, Jean-Christophe Simon, Alexandra Magro, Benoit Pujol, Etienne Danchin and Jean-Louis Hemptinne | <p>Trophic cascades, the indirect effect of predators on non-adjacent lower trophic levels, are important drivers of the structure and dynamics of ecological communities. However, the influence of intraspecific trait variation on the strength of t... |  | Community ecology, Eco-evolutionary dynamics, Food webs, Population ecology | Sara Magalhães | | 2019-08-02 09:11:03 | View |
A flexible pipeline combining clustering and correction tools for prokaryotic and eukaryotic metabarcoding
Miriam I Brandt, Blandine Trouche, Laure Quintric, Patrick Wincker, Julie Poulain, Sophie Arnaud-Haond
https://doi.org/10.1101/717355
A flexible pipeline combining clustering and correction tools for prokaryotic and eukaryotic metabarcoding
Recommended by Stefaniya Kamenova based on reviews by Tiago Pereira and 1 anonymous reviewer based on reviews by Tiago Pereira and 1 anonymous reviewer
High-throughput sequencing-based techniques such as DNA metabarcoding are increasingly advocated as providing numerous benefits over morphology‐based identifications for biodiversity inventories and ecosystem biomonitoring [1]. These benefits are particularly apparent for highly-diversified and/or hardly accessible aquatic and marine environments, where simple water or sediment samples could already produce acceptably accurate biodiversity estimates based on the environmental DNA present in the samples [2,3]. However, sequence-based characterization of biodiversity comes with its own challenges. A major one resides in the capacity to disentangle true biological diversity (be it taxonomic or genetic) from artefactual diversity generated by sequence-errors accumulation during PCR and sequencing processes, or from the amplification of non-target genes (i.e. pseudo-genes). On one hand, the stringent elimination of sequence variants might lead to biodiversity underestimation through the removal of true species, or the clustering of closely-related ones. On the other hand, a more permissive sequence filtering bears the risks of biodiversity inflation. Recent studies have outlined an excellent methodological framework for addressing this issue by proposing bioinformatic tools that allow the amplicon-specific error-correction as alternative or as complement to the more arbitrary approach of clustering into Molecular Taxonomic Units (MOTUs) based on sequence dissimilarity [4,5]. But to date, the relevance of amplicon-specific error-correction tools has been demonstrated only for a limited set of taxonomic groups and gene markers.
The study of Brandt et al. [6] successfully builds upon existing methodological frameworks for filling this gap in current literature. By proposing a bioinformatic pipeline combining Amplicon Sequence Variants (ASV) curation with MOTU clustering and additional post-clustering curation, the authors show that contrary to previous recommendations, ASV-based curation alone does not represent an adequate approach for DNA metabarcoding-based inventories of metazoans. Metazoans indeed, do exhibit inherently higher intra-specific and intra-individual genetic variability, necessarily leading to biased biodiversity estimates unbalanced in favor of species with higher intraspecific diversity in the absence of MOTU clustering. Interestingly, the positive effect of additional clustering showed to be dependent on the target gene region. Additional clustering had proportionally higher effect on the more polymorphic mitochondrial COI region (as compared to the 18S ribosomal gene). Thus, the major advantage of the study lies in the provision of optimal curation parameters that reflect the best possible balance between minimizing the impact of PCR/sequencing errors and the loss of true biodiversity across markers with contrasting levels of intragenomic variation. This is important as combining multiple markers is increasingly considered for improving the taxonomic coverage and resolution of data in DNA metabarcoding studies.
Another critical aspect of the study is the taxonomic assignation of curated OTUs (which is also the case for the majority of DNA metabarcoding-based biodiversity assessments). Facing the double challenge of focusing on taxonomic groups that are both highly diverse and poorly represented in public sequence reference databases, the authors failed to obtain high-resolution taxonomic assignments for several of the most closely-related species. As a result, taxa with low divergence levels were clustered as single taxonomic units, subsequently leading to underestimation of true biodiversity present. This finding adds to the argument that in order to be successful, sequence-based techniques still require the availability of comprehensive, high-quality reference databases.
Perhaps the only regret we might have with the study is the absence of mock community validation for the prokaryotes compartment. Even though the analyses of natural samples seem to suggest a positive effect of the curation pipeline, the concept of intra- versus inter-species variation in naturally occurring prokaryote communities remains at best ambiguous. Of course, constituting a representative sample of taxonomically-resolved prokaryote taxa from deep-sea habitats does not come without difficulties but has the benefit of opening opportunities for further studies on the matter.
References
[1] Porter, T. M., and Hajibabaei, M. (2018). Scaling up: A guide to high-throughput genomic approaches for biodiversity analysis. Molecular Ecology, 27(2), 313–338. doi: 10.1111/mec.14478
[2] Valentini, A., Taberlet, P., Miaud, C., Civade, R., Herder, J., Thomsen, P. F., … Dejean, T. (2016). Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Molecular Ecology, 25(4), 929–942. doi: 10.1111/mec.13428
[3] Leray, M., and Knowlton, N. (2015). DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proceedings of the National Academy of Sciences, 112(7), 2076–2081. doi: 10.1073/pnas.1424997112
[4] Callahan, B. J., McMurdie, P. J., and Holmes, S. P. (2017). Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. The ISME Journal, 11(12), 2639–2643. doi: 10.1038/ismej.2017.119
[5] Edgar, R. C. (2016). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv, 081257. doi: 10.1101/081257
[6] Brandt, M. I., Trouche, B., Quintric, L., Wincker, P., Poulain, J., and Arnaud-Haond, S. (2020). A flexible pipeline combining clustering and correction tools for prokaryotic and eukaryotic metabarcoding. BioRxiv, 717355, ver. 3 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/717355
| A flexible pipeline combining clustering and correction tools for prokaryotic and eukaryotic metabarcoding | Miriam I Brandt, Blandine Trouche, Laure Quintric, Patrick Wincker, Julie Poulain, Sophie Arnaud-Haond | <p>Environmental metabarcoding is an increasingly popular tool for studying biodiversity in marine and terrestrial biomes. With sequencing costs decreasing, multiple-marker metabarcoding, spanning several branches of the tree of life, is becoming ... | 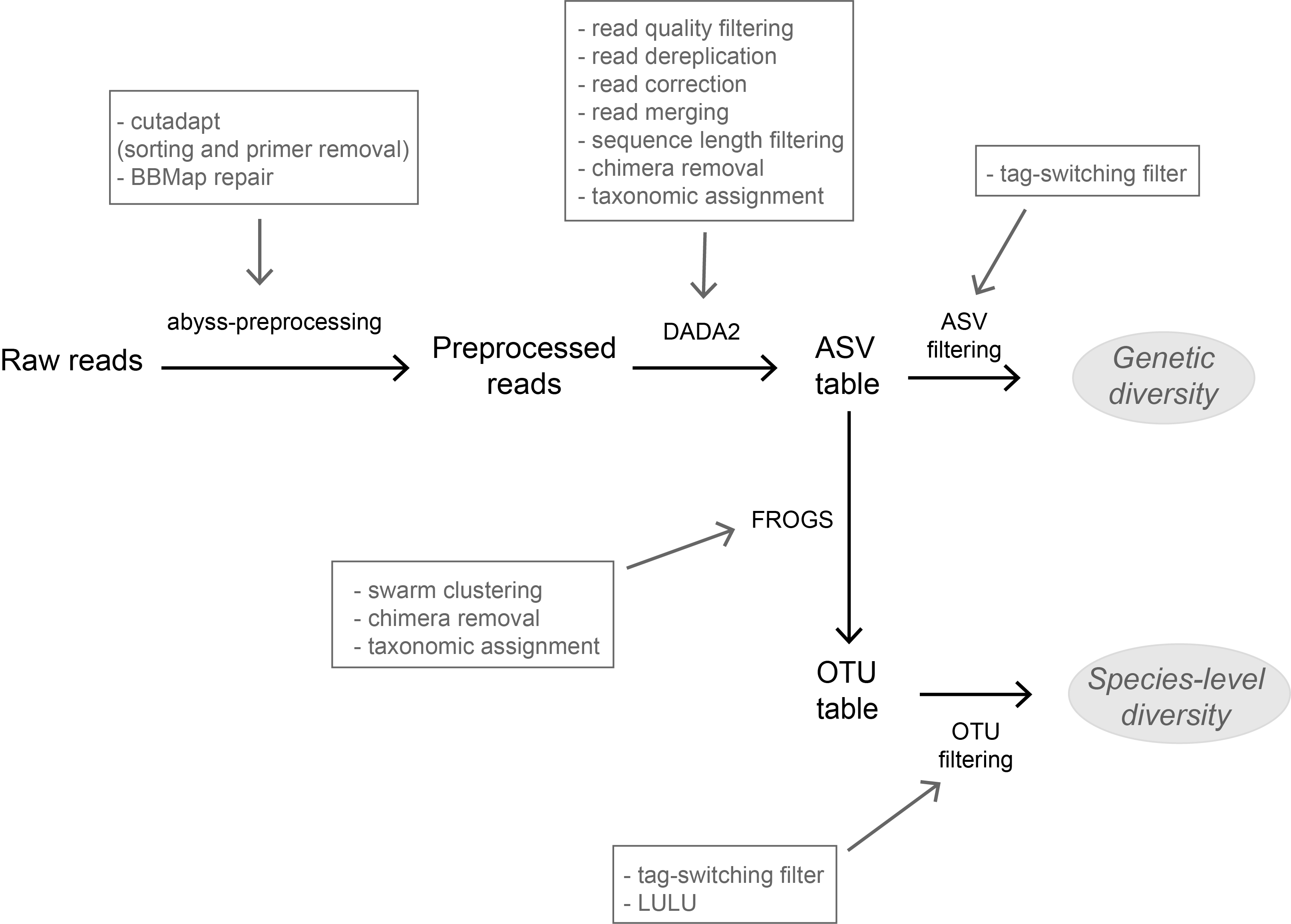 | Biodiversity, Community ecology, Marine ecology, Molecular ecology | Stefaniya Kamenova | | 2019-08-02 20:52:45 | View |
Are condition indices positively related to each other and to fitness?: a test with grackles
Recommended by Marcos Mendez based on reviews by Javier Seoane and Isabel López-Rull
Reproductive succes, as a surrogate of individual fitness, depends both on extrinsic and intrinsic factors [1]. Among the intrinsic factors, resource level or health are considered important potential drivers of fitness but exceedingly difficult to measure directly. Thus, a host of proxies have been suggested, known as condition indices [2]. The question arises whether all condition indices consistently measure the same "inner state" of individuals and whether all of them similarly correlate to individual fitness. In this preregistration, Berens and colleagues aim to answer this question for two common condition indices, fat score and scaled mass index (Fig. 1), using great-tailed grackles as a model system. Although this question is not new, it has not been satisfactorily solved and both reviewers found merit in the attempt to clarify this matter.

Figure 1. Hypothesized relationships between two condition indices and reproductive success. Single arrow heads indicate causal relationships; double arrow heads indicate only correlation. In a best case scenario, all relationships should be positive and linear.
A problem in adressing this question with grackles is limited population, ergo sample, size and limited possibilites of recapture individuals. Some relationships can be missed due to low statistical power. Unfortunately, existing tools for power analysis fall behind complex designs and the one planned for this study. Thus, any potentially non significant relationship has to be taken cautiously. Nevertheless, even if grackles will not provide a definitive answer (they never meant to do it), this preregistration can inspire broader explorations of matches and mismatches across condition indices and species, as well as uncover non-linear relationships with reproductive success.
References
[1] Roff, D. A. (2001). Life history evolution. Oxford University Press, Oxford.
[2] Labocha, M. K.; Hayes, J. P. (2012). Morphometric indices of body condition in birds: a review. Journal of Ornithology 153: 1–22. doi: 10.1007/s10336-011-0706-1
| Validating morphological condition indices and their relationship with reproductive success in great-tailed grackles | Jennifer M. Berens, Corina J. Logan, Melissa Folsom, Luisa Bergeron, Kelsey B. McCune | Morphological variation among individuals has the potential to influence multiple life history characteristics such as dispersal, migration, reproductive fitness, and survival (Wilder, Raubenheimer, and Simpson (2016)). Theoretically, individuals ... |  | Behaviour & Ethology, Conservation biology, Demography, Morphometrics, Preregistrations, Zoology | Marcos Mendez | | 2019-08-05 20:05:56 | View |
Stoichiometric constraints modulate the effects of temperature and nutrients on biomass distribution and community stability
Arnaud Sentis, Bart Haegeman, and José M. Montoya
https://doi.org/10.1101/589895
On the importance of stoichiometric constraints for understanding global change effects on food web dynamics
Recommended by Elisa Thebault based on reviews by 2 anonymous reviewers
The constraints associated with the mass balance of chemical elements (i.e. stoichiometric constraints) are critical to our understanding of ecological interactions, as outlined by the ecological stoichiometry theory [1]. Species in ecosystems differ in their elemental composition as well as in their level of elemental homeostasis [2], which can determine the outcome of interactions such as herbivory or decomposition on species coexistence and ecosystem functioning [3, 4].
Despite their importance, stoichiometric constraints are still often ignored in theoretical studies exploring the consequences of environmental perturbations on food web stability. Meanwhile, drivers of global change strongly alter biochemical cycles and the balance of chemical elements in ecosystems [5]. An important challenge is thus to understand how stoichiometric constraints affect food web responses to global changes.
The study of Sentis et al. [6] makes a step in that direction. This article investigates how stoichiometric constraints affect the response of consumer-resource dynamics to increasing temperature and nutrient inputs. It shows that the stoichiometric flexibility of the resource, coupled with lower consumer assimilation efficiency when stoichiometric unbalance between the resource and the consumer is higher, dampens the destabilizing effects of nutrient enrichment on species dynamics but reduces consumer persistence at extreme temperatures. Interestingly, these effects of stoichiometric constraints arise not only from changes in species assimilation efficiencies and carrying capacities but also from stoichiometric negative feedback loops on resource and consumer populations.
The results of this study are a call to further include stoichiometric constraints into food web models to better understand and predict the consequences of global changes on ecological communities. Many perspectives exist on that issue. For instance, it would be interesting to assess the effects of other stoichiometric mechanisms (e.g. changes in the element limiting growth [3]) on food web stability and its response to nutrient enrichment, as well as the effects of other global change drivers associated with altered biochemical cycles (e.g. carbon dioxide increase).
References
[1] Sterner, R. W. and Elser, J. J. (2017). Ecological Stoichiometry, The Biology of Elements from Molecules to the Biosphere. doi: 10.1515/9781400885695
[2] Elser, J. J., Sterner, R. W., Gorokhova, E., Fagan, W. F., Markow, T. A., Cotner, J. B., Harrison, J.F., Hobbie, S.E., Odell, G.M., Weider, L. W. (2000). Biological stoichiometry from genes to ecosystems. Ecology Letters, 3(6), 540–550. doi: 10.1111/j.1461-0248.2000.00185.x
[3] Daufresne, T., and Loreau, M. (2001). Plant–herbivore interactions and ecological stoichiometry: when do herbivores determine plant nutrient limitation? Ecology Letters, 4(3), 196–206. doi: 10.1046/j.1461-0248.2001.00210.x
[4] Zou, K., Thébault, E., Lacroix, G., and Barot, S. (2016). Interactions between the green and brown food web determine ecosystem functioning. Functional Ecology, 30(8), 1454–1465. doi: 10.1111/1365-2435.12626
[5] Peñuelas, J., Poulter, B., Sardans, J., Ciais, P., van der Velde, M., Bopp, L., Boucher, O., Godderis, Y., Hinsinger, P., Llusia, J., Nardin, E., Vicca, S., Obersteiner, M., Janssens, I. A. (2013). Human-induced nitrogen–phosphorus imbalances alter natural and managed ecosystems across the globe. Nature Communications, 4(1), 1–10. doi: 10.1038/ncomms3934
[6] Sentis, A., Haegeman, B. & Montoya, J.M. (2020). Stoichiometric constraints modulate the effects of temperature and nutrients on biomass distribution and community stability. bioRxiv, 589895, ver. 7 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/589895
| Stoichiometric constraints modulate the effects of temperature and nutrients on biomass distribution and community stability | Arnaud Sentis, Bart Haegeman, and José M. Montoya | <p>Temperature and nutrients are two of the most important drivers of global change. Both can modify the elemental composition (i.e. stoichiometry) of primary producers and consumers. Yet their combined effect on the stoichiometry, dynamics, and s... |  | Climate change, Community ecology, Food webs, Theoretical ecology, Thermal ecology | Elisa Thebault | | 2019-08-08 12:20:08 | View |
Eco-evolutionary dynamics further weakens mutualistic interaction and coexistence under population decline
Avril Weinbach, Nicolas Loeuille, Rudolf P. Rohr
https://doi.org/10.1101/570580
Doomed by your partner: when mutualistic interactions are like an evolutionary millstone around a species’ neck
Recommended by Sylvain Billiard based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Mutualistic interactions are the weird uncles of population and community ecology. They are everywhere, from the microbes aiding digestion in animals’ guts to animal-pollination services in ecosystems; They increase productivity through facilitation; They fascinate us when small birds pick the teeth of a big-mouthed crocodile. Yet, mutualistic interactions are far less studied and understood than competition or predation. Possibly because we are naively convinced that there is no mystery here: isn’t it obvious that mutualistic interactions necessarily facilitate species coexistence? Since mutualistic species benefit from one another, if one species evolves, the other should just follow, isn’t that so?
It is not as simple as that, for several reasons. First, because simple mutualistic Lotka-Volterra models showed that most of the time mutualistic systems should drift to infinity and be unstable (e.g. Goh 1979). This is not what happens in natural populations, so something is missing in simple models. At a larger scale, that of communities, this is even worse, since we are still far from understanding the link between the topology of mutualistic networks and the stability of a community. Second, interactions are context-dependent: mutualistic species exchange resources, and thus from the point of view of one species the interaction is either beneficial or not, depending on the net gain of energy (e.g. Holland and DeAngelis 2010). In other words, considering interactions as mutualistic per se is too caricatural. Third, since evolution is blind, the evolutionary response of a species to an environmental change can have any effect on its mutualistic partner, and not necessarily a neutral or positive effect. This latter reason is particularly highlighted by the paper by A. Weinbach et al. (2021).
Weinbach et al. considered a simple two-species mutualistic Lotka-Volterra model and analyzed the evolutionary dynamics of a trait controlling for the rate of interaction between the two species by using the classical Adaptive Dynamics framework. They showed that, depending on the form of the trade-off between this interaction trait and its effect on the intrinsic growth rate, several situations can occur at evolutionary equilibrium: species can stably coexist and maintain their interaction, or the interaction traits can evolve to zero where species can coexist without any interactions.
Weinbach et al. then investigated the fate of the two-species system if a partner species is strongly affected by environmental change, for instance, a large decrease of its growth rate. Because of the supposed trade-off between the interaction trait and the growth rate, the interaction trait in the focal species tends to decrease as an evolutionary response to the decline of the partner species. If environmental change is too large, the interaction trait can evolve to zero and can lead the partner species to extinction. An “evolutionary murder”.
Even though Weinbach et al. interpreted the results of their model through the lens of plant-pollinators systems, their model is not specific to this case. On the contrary, it is very general, which has advantages and caveats. By its generality, the model is informative because it is a proof of concept that the evolution of mutualistic interactions can have unexpected effects on any category of mutualistic systems. Yet, since the model lacks many specificities of plant-pollinator interactions, it is hard to evaluate how their result would apply to plant-pollinators communities.
I wanted to recommend this paper as a reminder that it is certainly worth studying the evolution of mutualistic interactions, because i) some unexpected phenomenons can occur, ii) we are certainly too naive about the evolution and ecology of mutualistic interactions, and iii) one can wonder to what extent we will be able to explain the stability of mutualistic communities without accounting for the co-evolutionary dynamics of mutualistic species.
References
Goh BS (1979) Stability in Models of Mutualism. The American Naturalist, 113, 261–275. http://www.jstor.org/stable/2460204.
Holland JN, DeAngelis DL (2010) A consumer–resource approach to the density-dependent population dynamics of mutualism. Ecology, 91, 1286–1295. https://doi.org/10.1890/09-1163.1
Weinbach A, Loeuille N, Rohr RP (2021) Eco-evolutionary dynamics further weakens mutualistic interaction and coexistence under population decline. bioRxiv, 570580, ver. 5 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/570580
| Eco-evolutionary dynamics further weakens mutualistic interaction and coexistence under population decline | Avril Weinbach, Nicolas Loeuille, Rudolf P. Rohr | <p style="text-align: justify;">With current environmental changes, evolution can rescue declining populations, but what happens to their interacting species? Mutualistic interactions can help species sustain each other when their environment wors... |  | Coexistence, Eco-evolutionary dynamics, Evolutionary ecology, Interaction networks, Pollination, Theoretical ecology | Sylvain Billiard | | 2019-09-05 11:29:45 | View |







 based on reviews by Sylvine Durand and 1 anonymous reviewer
based on reviews by Sylvine Durand and 1 anonymous reviewer




 based on reviews by Tiago Pereira and 1 anonymous reviewer
based on reviews by Tiago Pereira and 1 anonymous reviewer




 based on reviews by 2 anonymous reviewers
based on reviews by 2 anonymous reviewers












