Direct submissions to PCI Ecology from bioRxiv.org are possible using the B2J service
Latest recommendations

| Id | Title * | Authors * | Abstract * | Picture * ▼ | Thematic fields * | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
22 May 2019

Sex makes them sleepy: host reproductive status induces diapause in a parasitoid population experiencing harsh wintersTougeron K., Brodeur J., van Baaren J., Renault D. and Le Lann C. https://doi.org/10.1101/371385The response of interacting species to biotic seasonal cuesRecommended by Adele Mennerat and Enric Frago based on reviews by Anne Duplouy and 1 anonymous reviewerIn temperate regions, food abundance and quality vary greatly throughout the year, and the ability of organisms to synchronise their phenology to these changes is a key determinant of their reproductive success. Successful synchronisation requires that cues are perceived prior to change, leaving time for physiological adjustments. References [1] Tauber, M. J., Tauber, C. A., and Masaki, S. (1986). Seasonal Adaptations of Insects. Oxford, New York: Oxford University Press. | Sex makes them sleepy: host reproductive status induces diapause in a parasitoid population experiencing harsh winters | Tougeron K., Brodeur J., van Baaren J., Renault D. and Le Lann C. | <p>When organisms coevolve, any change in one species can induce phenotypic changes in traits and ecology of the other species. The role such interactions play in ecosystems is central, but their mechanistic bases remain underexplored. Upper troph... |  | Coexistence, Evolutionary ecology, Experimental ecology, Host-parasite interactions, Physiology | Adele Mennerat | 2018-07-18 18:51:03 | View | |
10 Jan 2024
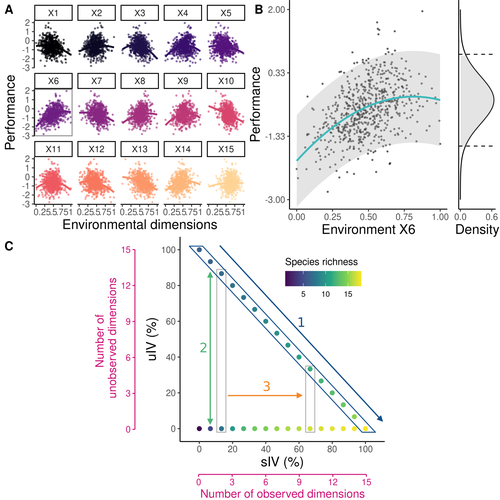
Beyond variance: simple random distributions are not a good proxy for intraspecific variability in systems with environmental structureCamille Girard-Tercieux, Ghislain Vieilledent, Adam Clark, James S. Clark, Benoit Courbaud, Claire Fortunel, Georges Kunstler, Raphaël Pélissier, Nadja Rüger, Isabelle Maréchaux https://doi.org/10.1101/2022.08.06.503032Two paradigms for intraspecific variabilityRecommended by Matthieu Barbier based on reviews by Simon Blanchet and Bart Haegeman based on reviews by Simon Blanchet and Bart Haegeman
Community ecology usually concerns itself with understanding the causes and consequences of diversity at a given taxonomic resolution, most classically at the species level. Yet there is no doubt that diversity exists at all scales, and phenotypic variability within a taxon can be comparable to differences between taxa, as observed from bacteria to fish and trees. The question that motivates an active and growing body of work (e.g. Raffard et al 2019) is not so much whether intraspecific variability matters, but what we get wrong by ignoring it and how to incorporate it into our understanding of communities. There is no established way to think about diversity at multiple nested taxonomic levels, and it is tempting to summarize intraspecific variability simply by measuring species mean and variance in any trait and metric. In this study, Girard-Tercieux et al (2023a) propose that, to understand its impact on community-level outcomes and in particular on species coexistence, we should carefully distinguish between two ways of thinking about intraspecific variability: -"unstructured" variation, where every individual's features are like an independent random draw from a species-specific distribution, for instance, due to genetic lottery and developmental accidents -"structured" variation that is due to each individual encountering a different but enduring microenvironment. The latter type of variability may still appear complex and random-like when the environment is high-dimensional (i.e. multifaceted, with many different factors contributing to each individual's performance and development). Thus, it is not necessarily "structured" in the sense of being easily understood -- we may need to measure more aspects of the environment than is practical if we want to fully predict these variations. What distinguishes this "structured" variability is that it is, in a loose sense, inheritable: individuals from the same species that grow in the same microenvironment will have the same performance, in a repeatable fashion. Thus, if each species is best at exploiting at least a fraction of environmental conditions, it is likely to avoid extinction by competition, except in the unlucky case of no propagule reaching any of the favorable sites. The core intuition, that the complex spatial structure and high-dimensional nature of the environment plays a key explanatory role in species coexistence, is a running thread through several of the authors' work (e.g. Clark et al 2010), clearly inspired by their focus on tropical forests. This study, by tackling the question of intraspecific determinants of interspecific outcomes, makes a compelling addition to this line of investigation, coming as a theoretical companion to a more data-oriented study (Girard-Tercieux et al 2023b). But I believe it raises a question that is even broader in scope. This kind of intraspecific variability, due to different individuals growing in different microenvironments, is perhaps most relevant for trees and other sessile organisms, but the distinction made here between "unstructured" and "structured" variability can likely be extended to many other ecological settings. In my understanding, what matters most in "structured" variability is not so much it stemming from a fixed environment, but rather it being maintained across generations, rather than possibly lost by drift. This difference between variability in the form of "frozen" randomness and in the form of stochastic drift over time is highly relevant in other theoretical fields (e.g. in physics, where it is the difference between a disordered solid and a liquid), and thus, I expect that it is a meaningful distinction to make throughout community ecology. References James S. Clark, David Bell, Chengjin Chu, Benoit Courbaud, Michael Dietze, Michelle Hersh, Janneke HilleRisLambers et al. (2010) "High‐dimensional coexistence based on individual variation: a synthesis of evidence." Ecological Monographs 80, no. 4 : 569-608. https://doi.org/10.1890/09-1541.1 Camille Girard-Tercieux, Ghislain Vieilledent, Adam Clark, James S. Clark, Benoît Courbaud, Claire Fortunel, Georges Kunstler, Raphaël Pélissier, Nadja Rüger, Isabelle Maréchaux (2023a) "Beyond variance: simple random distributions are not a good proxy for intraspecific variability in systems with environmental structure." bioRxiv, ver. 4 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2022.08.06.503032 Camille Girard‐Tercieux, Isabelle Maréchaux, Adam T. Clark, James S. Clark, Benoît Courbaud, Claire Fortunel, Joannès Guillemot et al. (2023b) "Rethinking the nature of intraspecific variability and its consequences on species coexistence." Ecology and Evolution 13, no. 3 : e9860. https://doi.org/10.1002/ece3.9860 Allan Raffard, Frédéric Santoul, Julien Cucherousset, and Simon Blanchet. (2019) "The community and ecosystem consequences of intraspecific diversity: A meta‐analysis." Biological Reviews 94, no. 2: 648-661. https://doi.org/10.1111/brv.12472 | Beyond variance: simple random distributions are not a good proxy for intraspecific variability in systems with environmental structure | Camille Girard-Tercieux, Ghislain Vieilledent, Adam Clark, James S. Clark, Benoit Courbaud, Claire Fortunel, Georges Kunstler, Raphaël Pélissier, Nadja Rüger, Isabelle Maréchaux | <p>The role of intraspecific variability (IV) in shaping community dynamics and species coexistence has been intensively discussed over the past decade and modelling studies have played an important role in that respect. However, these studies oft... |  | Biodiversity, Coexistence, Community ecology, Competition, Theoretical ecology | Matthieu Barbier | 2022-08-07 12:51:30 | View | |
03 Oct 2023
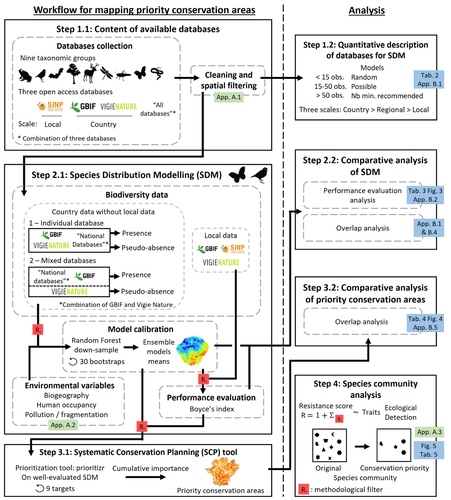
Integrating biodiversity assessments into local conservation planning: the importance of assessing suitable data sourcesThibaut Ferraille, Christian Kerbiriou, Charlotte Bigard, Fabien Claireau, John D. Thompson https://doi.org/10.1101/2023.05.09.539999Biodiversity databases are ever more numerous, but can they be used reliably for Species Distribution Modelling?Recommended by Nicolas Schtickzelle based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Proposing efficient guidelines for biodiversity conservation often requires the use of forecasting tools. Species Distribution Models (SDM) are more and more used to predict how the distribution of a species will react to environmental change, including any large-scale management actions that could be implemented. Their use is also boosted by the increase of publicly available biodiversity databases[1]. The now famous aphorism by George Box "All models are wrong but some are useful"[2] very well summarizes that the outcome of a model must be adjusted to, and will depend on, the data that are used to parameterize it. The question of the reliability of using biodiversity databases to parameterize biodiversity models such as SDM –but the question would also apply to other kinds of biodiversity models, e.g. Population Viability Analysis models[3]– is key to determine the confidence that can be placed in model predictions. This point is often overlooked by some categories of biodiversity conservation stakeholders, in particular the fact that some data were collected using controlled protocols while others are opportunistic. In this study[4], the authors use a collection of databases covering a range of species as well as of geographic scales in France and using different data collection and validation approaches as a case study to evaluate the impact of data quality when performing Strategic Environmental Assessment (SEA). Among their conclusions, the fact that a large-scale database (what they call the “country” level) is necessary to reliably parameterize SDM. Besides this and other conclusions of their study, which are likely to be in part specific to their case study –unfortunately for its conservation, biodiversity is complex and varies a lot–, the merit of this work lies in the approach used to test the impact of data on model predictions. References 1. Feng, X. et al. A review of the heterogeneous landscape of biodiversity databases: Opportunities and challenges for a synthesized biodiversity knowledge base. Global Ecology and Biogeography 31, 1242–1260 (2022). https://doi.org/10.1111/geb.13497 2. Box, G. E. P. Robustness in the Strategy of Scientific Model Building. in Robustness in Statistics (eds. Launer, R. L. & Wilkinson, G. N.) 201–236 (Academic Press, 1979). https://doi.org/10.1016/B978-0-12-438150-6.50018-2. 3. Beissinger, S. R. & McCullough, D. R. Population Viability Analysis. (The University of Chicago Press, 2002). 4. Ferraille, T., Kerbiriou, C., Bigard, C., Claireau, F. & Thompson, J. D. (2023) Integrating biodiversity assessments into local conservation planning: the importance of assessing suitable data sources. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.05.09.539999 | Integrating biodiversity assessments into local conservation planning: the importance of assessing suitable data sources | Thibaut Ferraille, Christian Kerbiriou, Charlotte Bigard, Fabien Claireau, John D. Thompson | <p>Strategic Environmental Assessment (SEA) of land-use planning is a fundamental tool to minimize environmental impacts of artificialization. In this context, Systematic Conservation Planning (SCP) tools based on Species Distribution Models (SDM)... |  | Biodiversity, Conservation biology, Species distributions, Terrestrial ecology | Nicolas Schtickzelle | 2023-05-11 09:41:05 | View | |
02 Aug 2022

The effect of dominance rank on female reproductive success in social mammalsShivani, Elise Huchard, Dieter Lukas https://doi.org/10.32942/osf.io/rc8naWhen do dominant females have higher breeding success than subordinates? A meta-analysis across social mammals.Recommended by Matthieu Paquet based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
In this meta-analysis, Shivani et al. [1] investigate 1) whether dominance and reproductive success are generally associated across social mammals and 2) whether this relationship varies according to a) life history traits (e.g., stronger for species with large litter size), b) ecological conditions (e.g., stronger when resources are limited) and c) the social environment (e.g., stronger for cooperative breeders than for plural breeders). Generally, the results are consistent with their predictions, except there was no clear support for this relationship to be conditional on the ecological conditions. considered As I have previously recommended the preregistration of this study [2,3], I do not have much to add here, as such recommendation should not depend on the outcome of the study. What I would like to recommend is the whole scientific process performed by the authors, from preregistration sent for peer review, to preprint submission and post-study peer review. It is particularly recommendable to notice that this project was a Masters student project, which shows that it is possible and worthy to preregister studies, even for such rather short-term projects. I strongly congratulate the authors for choosing this process even for an early career short-term project. I think it should be made possible for short-term students to conduct a preregistration study as a research project, without having to present post-study results. I hope this study can encourage a shift in the way we sometimes evaluate students’ projects. I also recommend the readers to look into the whole pre- and post- study reviewing history of this manuscript and the associated preregistration, as it provides a better understanding of the process and a good example of the associated challenges and benefits [4]. It was a really enriching experience and I encourage others to submit and review preregistrations and registered reports!
References [1] Shivani, Huchard, E., Lukas, D. (2022). The effect of dominance rank on female reproductive success in social mammals. EcoEvoRxiv, rc8na, ver. 10 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.32942/osf.io/rc8na [2] Shivani, Huchard, E., Lukas, D. (2020). Preregistration - The effect of dominance rank on female reproductive success in social mammals In principle acceptance by PCI Ecology of the version 1.2 on 07 July 2020. https://dieterlukas.github.io/Preregistration_MetaAnalysis_RankSuccess.html [3] Paquet, M. (2020) Why are dominant females not always showing higher reproductive success? A preregistration of a meta-analysis on social mammals. Peer Community in Ecology, 100056. https://doi.org/10.24072/pci.ecology.100056 [4] Parker, T., Fraser, H., & Nakagawa, S. (2019). Making conservation science more reliable with preregistration and registered reports. Conservation Biology, 33(4), 747-750. https://doi.org/10.1111/cobi.13342 | The effect of dominance rank on female reproductive success in social mammals | Shivani, Elise Huchard, Dieter Lukas | <p>Life in social groups, while potentially providing social benefits, inevitably leads to conflict among group members. In many social mammals, such conflicts lead to the formation of dominance hierarchies, where high-ranking individuals consiste... |  | Behaviour & Ethology, Meta-analyses | Matthieu Paquet | 2021-10-13 18:26:42 | View | |
31 Aug 2023
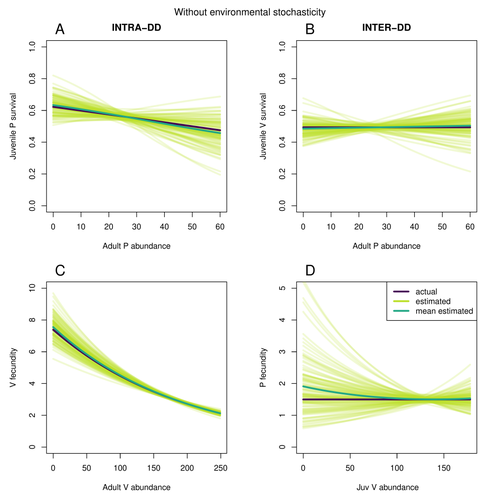
Assessing species interactions using integrated predator-prey modelsMatthieu Paquet, Frederic Barraquand https://doi.org/10.32942/X2RC7WAddressing the daunting challenge of estimating species interactions from count dataRecommended by Tim Coulson and David Alonso and David Alonso based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Trophic interactions are at the heart of community ecology. Herbivores consume plants, predators consume herbivores, and pathogens and parasites infect, and sometimes kill, individuals of all species in a food web. Given the ubiquity of trophic interactions, it is no surprise that ecologists and evolutionary biologists strive to accurately characterize them. The outcome of an interaction between individuals of different species depends upon numerous factors such as the age, sex, and even phenotype of the individuals involved and the environment in which they are in. Despite this complexity, biologists often simplify an interaction down to a single number, an interaction coefficient that describes the average outcome of interactions between members of the populations of the species. Models of interacting species tend to be very simple, and interaction coefficients are often estimated from time series of population sizes of interacting species. Although biologists have long known that this approach is often approximate and sometimes unsatisfactory, work on estimating interaction strengths in more complex scenarios, and using ecological data beyond estimates of abundance, is still in its infancy. In their paper, Matthieu Paquet and Frederic Barraquand (2023) develop a demographic model of a predator and its prey. They then simulate demographic datasets that are typical of those collected by ecologists and use integrated population modelling to explore whether they can accurately retrieve the values interaction coefficients included in their model. They show that they can with good precision and accuracy. The work takes an important step in showing that accurate interaction coefficients can be estimated from the types of individual-based data that field biologists routinely collect, and it paves for future work in this area. As if often the case with exciting papers such as this, the work opens up a number of other avenues for future research. What happens as we move from demographic models of two species interacting such as those used by Paquet and Barraquand to more realistic scenarios including multiple species? How robust is the approach to incorrectly specified process or observation models, core components of integrated population modelling that require detailed knowledge of the system under study? Integrated population models have become a powerful and widely used tool in single-species population ecology. It is high time the techniques are extended to community ecology, and this work takes an important step in showing that this should and can be done. I would hope the paper is widely read and cited. References Paquet, M., & Barraquand, F. (2023). Assessing species interactions using integrated predator-prey models. EcoEvoRxiv, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.32942/X2RC7W | Assessing species interactions using integrated predator-prey models | Matthieu Paquet, Frederic Barraquand | <p style="text-align: justify;">Inferring the strength of species interactions from demographic data is a challenging task. The Integrated Population Modelling (IPM) approach, bringing together population counts, capture-recapture, and individual-... |  | Community ecology, Demography, Euring Conference, Food webs, Population ecology, Statistical ecology | Tim Coulson | Ilhan Özgen-Xian | 2023-01-05 17:02:22 | View |
06 Oct 2020

Does space use behavior relate to exploration in a species that is rapidly expanding its geographic range?Kelsey B. McCune, Cody Ross, Melissa Folsom, Luisa Bergeron, Corina Logan http://corinalogan.com/Preregistrations/gspaceuse.htmlExplore and move: a key to success in a changing world?Recommended by Blandine Doligez based on reviews by Joe Nocera, Marion Nicolaus and Laure CauchardChanges in the spatial range of many species are one of the major consequences of the profound alteration of environmental conditions due to human activities. Some species expand, sometimes spectacularly during invasions; others decline; some shift. Because these changes result in local biodiversity loss (whether local species go extinct or are replaced by colonizing ones), understanding the factors driving spatial range dynamics appears crucial to predict biodiversity dynamics. Identifying the factors that shape individual movement is a main step towards such understanding. The study described in this preregistration (McCune et al. 2020) falls within this context by testing possible links between individual exploration behaviour and movements related to daily space use in an avian study model currently rapidly expanding, the great-tailed grackle (Quiscalus mexicanus). Movement and exploration: which direction(s) for the link between exploration and dispersal? Evolutionary and conservation perspectives References Badayev, A. V., Martin, T. E and Etges, W. J. 1996. Habitat sampling and habitat selection by female wild turkeys: ecological correlates and reproductive consequences. Auk 113: 636-646. doi: https://doi.org/10.2307/4088984 | Does space use behavior relate to exploration in a species that is rapidly expanding its geographic range? | Kelsey B. McCune, Cody Ross, Melissa Folsom, Luisa Bergeron, Corina Logan | Great-tailed grackles (Quiscalus mexicanus) are rapidly expanding their geographic range (Wehtje 2003). Range expansion could be facilitated by consistent behavioural differences between individuals on the range edge and those in other parts of th... |  | Behaviour & Ethology, Biological invasions, Conservation biology, Habitat selection, Phenotypic plasticity, Preregistrations, Spatial ecology, Metacommunities & Metapopulations | Blandine Doligez | 2019-09-30 19:27:40 | View | |
24 May 2024
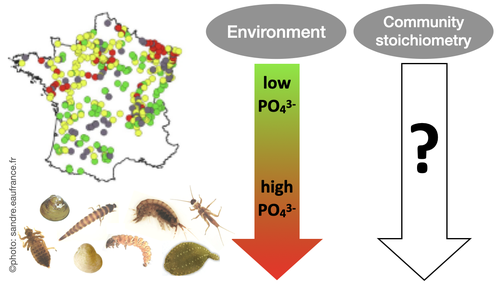
Effects of water nutrient concentrations on stream macroinvertebrate community stoichiometry: a large-scale studyMiriam Beck, Elise Billoir, Philippe Usseglio-Polatera, Albin Meyer, Edwige Gautreau, Michael Danger https://doi.org/10.1101/2024.02.01.574823The influence of water phosphorus and nitrogen loads on stream macroinvertebrate community stoichiometryRecommended by Huihuang Chen based on reviews by Thomas Guillemaud, Jun Zuo and 1 anonymous reviewer based on reviews by Thomas Guillemaud, Jun Zuo and 1 anonymous reviewer
The manuscript by Beck et al. (2024) investigates the effects of water phosphorus and nitrogen loads on stream macroinvertebrate community stoichiometry across France. Utilizing data from over 1300 standardized sampling events, this research finds that community stoichiometry is significantly influenced by water phosphorus concentration, with the strongest effects at low nitrogen levels. The results demonstrate that the assumptions of Ecological Stoichiometry Theory apply at the community level for at least two dominant taxa and across a broad spatial scale, with probable implications for nutrient cycling and ecosystem functionality. This manuscript contributes to ecological theory, particularly by extending Ecological Stoichiometry Theory to include community-level interactions, clarifying the impact of nutrient concentrations on community structure and function, and informing nutrient management and conservation strategies. In summary, this study not only addresses a gap in community-level stoichiometric research but also delivers crucial empirical support for advancing ecological science and promoting environmental stewardship. References Beck M, Billoir E, Usseglio-Polatera P, Meyer A, Gautreau E and Danger M (2024) Effects of water nutrient concentrations on stream macroinvertebrate community stoichiometry: a large-scale study. bioRxiv, 2024.02.01.574823, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2024.02.01.574823 | Effects of water nutrient concentrations on stream macroinvertebrate community stoichiometry: a large-scale study | Miriam Beck, Elise Billoir, Philippe Usseglio-Polatera, Albin Meyer, Edwige Gautreau, Michael Danger | <p>Basal resources generally mirror environmental nutrient concentrations in the elemental composition of their tissue, meaning that nutrient alterations can directly reach consumer level. An increased nutrient content (e.g. phosphorus) in primary... |  | Community ecology, Ecological stoichiometry | Huihuang Chen | Thomas Guillemaud, Jun Zuo, Anonymous | 2024-02-02 10:14:01 | View |
30 May 2024
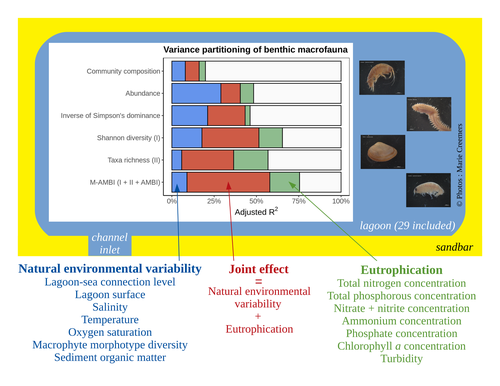
Disentangling the effects of eutrophication and natural variability on macrobenthic communities across French coastal lagoonsAuriane G. Jones, Gauthier Schaal, Aurélien Boyé, Marie Creemers, Valérie Derolez, Nicolas Desroy, Annie Fiandrino, Théophile L. Mouton, Monique Simier, Niamh Smith, Vincent Ouisse https://doi.org/10.1101/2022.08.18.504439Untangling Eutrophication Effects on Coastal Lagoon EcosystemsRecommended by Nathalie Niquil based on reviews by Kaylee P. Smit, Matthew J. Pruden and Kendyl Wright based on reviews by Kaylee P. Smit, Matthew J. Pruden and Kendyl Wright
Disentangling the effects on ecosystem structure and functioning of natural and human-induced impacts in transitional waters is a great challenge in coast ecology. This is due to the observation that the ecosystems of transitional waters are naturally dynamic systems with characteristics of stressed systems. For example, the benthic communities present low species richness and high abundance of species with a high tolerance to variations, e.g., salinity. This general observation is known as the paradigm of the “Transitional Waters Quality Paradox” (Zaldívar et al., 2008) derived from the previously described “Estuarine Quality Paradox” (Elliott and Quintino, 2007). In Jones et al. (2024) “Disentangling the effects of eutrophication and natural variability on macrobenthic communities across French coastal lagoons”, a great diversity of lagoons is analyzed to disentangle the effects of eutrophication from those of natural environmental variability on benthic macroinvertebrates and understanding the links between environmental variables affecting benthic macroinvertebrates. These authors use a very elegant set of numerical approaches, including correlograms, linear models and variance partitioning. They apply this suite to a dataset of macrobenthic invertebrate abundances and environmental variables from 29 Mediterranean coastal lagoons in France. Through this suite of analyses, they demonstrate the strong complexity of the mechanisms interplaying in a situation of eutrophication on lagoon macrobenthos. The mechanisms involved are direct, like toxicity, or indirect, for example, through modifications of the sediment's biogeochemistry. Such a result on the different interactions involved is very important in the context of the search for indicators to define ecosystem status. Improving the definition of metrics is essential in environmental management decisions. References Elliott, M. and Quintino, V. (2007) The estuarine quality paradox, environmental homeostasis and the difficulty of detecting anthropogenic stress in naturally stressed areas. Marine Pollution Bulletin 54, 640–645. https://doi.org/10.1016/j.marpolbul.2007.02.003 Zaldívar, J. (2008). Eutrophication in transitional waters: an overview. https://doi.org/10.1285/I18252273V2N1P1 | Disentangling the effects of eutrophication and natural variability on macrobenthic communities across French coastal lagoons | Auriane G. Jones, Gauthier Schaal, Aurélien Boyé, Marie Creemers, Valérie Derolez, Nicolas Desroy, Annie Fiandrino, Théophile L. Mouton, Monique Simier, Niamh Smith, Vincent Ouisse | <p style="text-align: justify;">Coastal lagoons are transitional ecosystems that host a unique diversity of species and support many ecosystem services. Owing to their position at the interface between land and sea, they are also subject to increa... |  | Biodiversity, Community ecology, Ecosystem functioning, Marine ecology | Nathalie Niquil | Matthew J. Pruden | 2023-09-08 11:26:01 | View |
10 Oct 2018

Detecting within-host interactions using genotype combination prevalence dataSamuel Alizon, Carmen Lía Murall, Emma Saulnier, Mircea T Sofonea https://doi.org/10.1101/256586Combining epidemiological models with statistical inference can detect parasite interactionsRecommended by Dustin Brisson based on reviews by Samuel Díaz Muñoz, Erick Gagne and 1 anonymous reviewerThere are several important topics in the study of infectious diseases that have not been well explored due to technical difficulties. One such topic is pursued by Alizon et al. in “Modelling coinfections to detect within-host interactions from genotype combination prevalences” [1]. Both theory and several important examples have demonstrated that interactions among co-infecting strains can have outsized impacts on disease outcomes, transmission dynamics, and epidemiology. Unfortunately, empirical data on pathogen interactions and their outcomes is often correlational making results difficult to decipher. References [1] Alizon, S., Murall, C.L., Saulnier, E., & Sofonea, M.T. (2018). Detecting within-host interactions using genotype combination prevalence data. bioRxiv, 256586, ver. 3 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/256586 | Detecting within-host interactions using genotype combination prevalence data | Samuel Alizon, Carmen Lía Murall, Emma Saulnier, Mircea T Sofonea | <p>Parasite genetic diversity can provide information on disease transmission dynamics but most methods ignore the exact combinations of genotypes in infections. We introduce and validate a new method that combines explicit epidemiological modelli... |  | Eco-immunology & Immunity, Epidemiology, Host-parasite interactions, Statistical ecology | Dustin Brisson | Samuel Díaz Muñoz, Erick Gagne | 2018-02-01 09:23:26 | View |
23 Mar 2020

Intraspecific difference among herbivore lineages and their host-plant specialization drive the strength of trophic cascadesArnaud Sentis, Raphaël Bertram, Nathalie Dardenne, Jean-Christophe Simon, Alexandra Magro, Benoit Pujol, Etienne Danchin and Jean-Louis Hemptinne https://doi.org/10.1101/722140Tell me what you’ve eaten, I’ll tell you how much you’ll eat (and be eaten)Recommended by Sara Magalhães and Raul Costa-Pereira based on reviews by Bastien Castagneyrol and 1 anonymous reviewerTritrophic interactions have a central role in ecological theory and applications [1-3]. Particularly, systems comprised of plants, herbivores and predators have historically received wide attention given their ubiquity and economic importance [4]. Although ecologists have long aimed to understand the forces that govern alternating ecological effects at successive trophic levels [5], several key open questions remain (at least partially) unanswered [6]. In particular, the analysis of complex food webs has questioned whether ecosystems can be viewed as a series of trophic chains [7,8]. Moreover, whether systems are mostly controlled by top-down (trophic cascades) or bottom-up processes remains an open question [6]. References [1] Price, P. W., Bouton, C. E., Gross, P., McPheron, B. A., Thompson, J. N., & Weis, A. E. (1980). Interactions among three trophic levels: influence of plants on interactions between insect herbivores and natural enemies. Annual review of Ecology and Systematics, 11(1), 41-65. doi: 10.1146/annurev.es.11.110180.000353 | Intraspecific difference among herbivore lineages and their host-plant specialization drive the strength of trophic cascades | Arnaud Sentis, Raphaël Bertram, Nathalie Dardenne, Jean-Christophe Simon, Alexandra Magro, Benoit Pujol, Etienne Danchin and Jean-Louis Hemptinne | <p>Trophic cascades, the indirect effect of predators on non-adjacent lower trophic levels, are important drivers of the structure and dynamics of ecological communities. However, the influence of intraspecific trait variation on the strength of t... |  | Community ecology, Eco-evolutionary dynamics, Food webs, Population ecology | Sara Magalhães | 2019-08-02 09:11:03 | View |
MANAGING BOARD
Julia Astegiano
Tim Coulson
Anna Eklof
Dominique Gravel
François Massol
Ben Phillips
Cyrille Violle










