
SCHTICKZELLE Nicolas
- Earth and Life Institute - Biodiversity Research Centre, Université catholique de Louvain, Louvain-la-Neuve, Belgium
- Climate change, Conservation biology, Demography, Dispersal & Migration, Experimental ecology, Phenotypic plasticity, Population ecology, Spatial ecology, Metacommunities & Metapopulations, Statistical ecology
- recommender
Recommendations: 3
Reviews: 0
Recommendations: 3
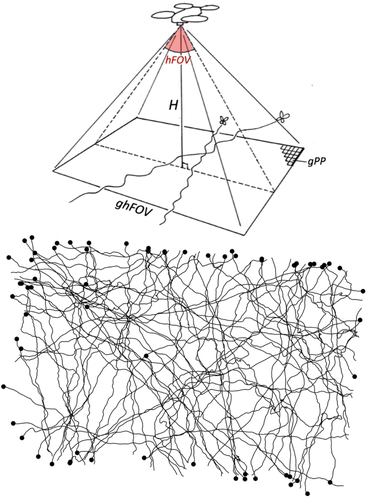
Tracking butterfly flight in the field from an unmanned aerial vehicle (UAV): a methodological proof of principle.
Breaking barriers in butterfly tracking: how drone technology and image analysis could boost movement ecology of butterflies
Recommended by Nicolas Schtickzelle based on reviews by 3 anonymous reviewersUnderstanding how animals move within and across landscapes is fundamental to behavioural ecology, conservation biology, and movement ecology. Tracking movement provides insights into migration and dispersal patterns, habitat preferences, intra- and interspecific interactions, etc. For long, movement recording was limited to indirect methods, such as Capture-Mark-Recapture. Despite being at the basis of an incredible amount of knowledge and developments in ecology, these methods do not inform on the movement path itself, just its beginning and end. Tracking individuals during their movement was really needed.
Over the years, researchers have developed a range of tracking methodologies, with technological innovations continually improving precision and efficiency (Trappes, 2023). While tracking large terrestrial and marine animals and birds is now well-established using GPS telemetry and biologging, monitoring small flying insects remains a significant challenge due to their size, erratic flight patterns, and sensitivity to environmental disturbances. It is especially the case for butterflies due to their lightweight bodies and relative low flight power. Given the role butterflies play as model organisms in diverse areas of ecology, research to allow tracking their movement path is of prime interest.
I remember the many hours I spent, in a time (early 2000s) GPS technology was still quite imprecise, following butterflies for a distance, placing sticks at turning points and reconstructing afterwards the movement path by triangulating the distances of sticks to know location marks (Schtickzelle et al., 2007). It was quite effective but prohibitive in terms of resources. Later came GPS devices precise enough for an individual to run in the footsteps of a butterfly to record its path. Still, methods have been highly desirable that could track butterflies with some level of automation and from a distance. Experiments were performed with harmonic radar (attaching a passive transponder that reflects radar signals; Cant et al., 2005) but were never largely adopted given they required acquiring and positioning costly and heavy equipment and maintaining at all time a direct line of sight with the tracked butterfly.
Here comes this pioneering study by de Margerie and Monmasson (Margerie & Monmasson, 2025) who introduce an innovative approach using a consumer-level commercial drone to track butterfly flight, offering a promising solution for long-duration, high-resolution flight trajectory analysis in natural habitats. Their study is a proof of principle that a drone, hovering in a fixed position, can be used as a flying platform to capture high-resolution vertical imagery to precisely record butterfly flight movements. Images are then analysed to reconstruct the flight path, a point on which they developed innovative approaches in the study.
The study therefore represents a significant leap forward in butterfly flight tracking methodology, with technology that many labs could acquire and operate. Further research is needed to alleviate some of the current limitations before large-scale adoption to track butterfly movements in the field is within reach: e.g. the need for a very high contrast between the butterfly and the vegetation above which it flies (here white Pieris butterflies over a relatively homogeneous green crop field were filmed), the limits in spatiotemporal scale due to the fixed drone position and its short battery life, and some difficulties in image processing to reconstruct movement paths, in particular when several individuals would cross paths. Considering the fast progress in both the drone technology and image analysis techniques, such progress could however come faster than we might anticipate.
References
Cant E. T., Smith A. D., Reynolds D. R. & Osborne J. L. (2005). Tracking butterfly flight paths across the landscape with harmonic radar. Proceedings of the Royal Society of London B 272, 785–790. https://doi.org/10.1098/rspb.2004.3002
de Margerie E. & Monmasson K. (2025) Tracking butterfly flight in the field from an unmanned aerial vehicle (UAV): a methodological proof of principle. bioRxiv, ver.5 peer-reviewed and recommended by PCI Ecology https://doi.org/10.1101/2024.07.17.603869
Schtickzelle N., Joiris A., Van Dyck H., & Baguette M. (2007). Quantitative analysis of changes in movement behaviour within and outside habitat in a specialist butterfly. BMC Evolutionary Biology 7, 4. https://doi.org/10.1186/1471-2148-7-4
Trappes R. (2023). How tracking technology is transforming animal ecology: Epistemic values, interdisciplinarity, and technology-driven scientific change. Synthese 201, 128. https://doi.org/10.1007/s11229-023-04122-5
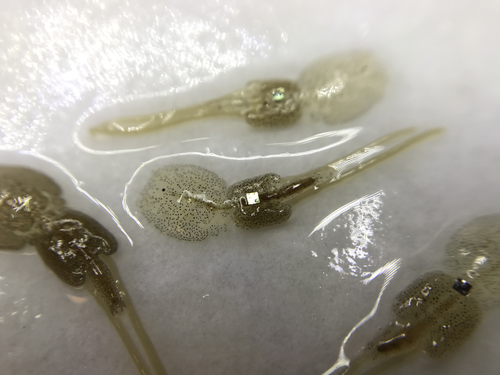
Methods for tagging an ectoparasite, the salmon louse Lepeophtheirus salmonis
Marking invertebrates using RFID tags
Recommended by Nicolas Schtickzelle based on reviews by Simon Blanchet and 1 anonymous reviewerGuiding and monitoring the efficiency of conservation efforts needs robust scientific background information, of which one key element is estimating wildlife abundance and its spatial and temporal variation. As raw counts are by nature incomplete counts of a population, correcting for detectability is required (Clobert, 1995; Turlure et al., 2018). This can be done with Capture-Mark-Recapture protocols (Iijima, 2020). Techniques for marking individuals are diverse, e.g. writing on butterfly wings, banding birds, or using natural specific patterns in the individual’s body such as leopard fur or whale tail. Advancement in technology opens new opportunities for developing marking techniques, including strategies to limit mark identification errors (Burchill & Pavlic, 2019), and for using active marks that can transmit data remotely or be read automatically.
The details of such methodological developments frequently remain unpublished, the method being briefly described in studies that use it. For a few years, there has been however a renewed interest in proper publishing of methods for ecology and evolution. This study by Folk & Mennerat (2023) fits in this context, offering a nice example of detailed description and testing of a method to mark salmon ectoparasites using RFID tags. Such tags are extremely small, yet easy to use, even with automatic recording procedure. The study provides a very good basis protocol that should help researchers working for small species, in particular invertebrates. The study is complemented by a video illustrating the placement of the tag so the reader who would like to replicate the procedure can get a very precise idea of it.
References
Burchill, A. T., & Pavlic, T. P. (2019). Dude, where’s my mark? Creating robust animal identification schemes informed by communication theory. Animal Behaviour, 154, 203–208. https://doi.org/10.1016/j.anbehav.2019.05.013
Clobert, J. (1995). Capture-recapture and evolutionary ecology: A difficult wedding ? Journal of Applied Statistics, 22(5–6), 989–1008.
Folk, A., & Mennerat, A. (2023). Methods for tagging an ectoparasite, the salmon louse Lepeophtheirus salmonis (p. 2023.08.31.555695). bioRxiv, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.08.31.555695
Iijima, H. (2020). A Review of Wildlife Abundance Estimation Models: Comparison of Models for Correct Application. Mammal Study, 45(3), 177–188. https://doi.org/10.3106/ms2019-0082
Turlure, C., Pe’er, G., Baguette, M., & Schtickzelle, N. (2018). A simplified mark–release–recapture protocol to improve the cost effectiveness of repeated population size quantification. Methods in Ecology and Evolution, 9(3), 645–656. https://doi.org/10.1111/2041-210X.12900
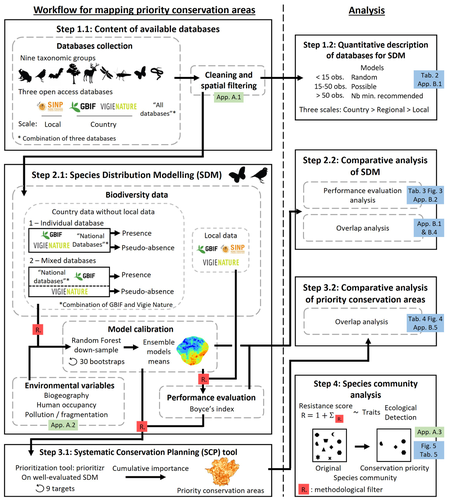
Integrating biodiversity assessments into local conservation planning: the importance of assessing suitable data sources
Biodiversity databases are ever more numerous, but can they be used reliably for Species Distribution Modelling?
Recommended by Nicolas Schtickzelle based on reviews by 2 anonymous reviewersProposing efficient guidelines for biodiversity conservation often requires the use of forecasting tools. Species Distribution Models (SDM) are more and more used to predict how the distribution of a species will react to environmental change, including any large-scale management actions that could be implemented. Their use is also boosted by the increase of publicly available biodiversity databases[1]. The now famous aphorism by George Box "All models are wrong but some are useful"[2] very well summarizes that the outcome of a model must be adjusted to, and will depend on, the data that are used to parameterize it. The question of the reliability of using biodiversity databases to parameterize biodiversity models such as SDM –but the question would also apply to other kinds of biodiversity models, e.g. Population Viability Analysis models[3]– is key to determine the confidence that can be placed in model predictions. This point is often overlooked by some categories of biodiversity conservation stakeholders, in particular the fact that some data were collected using controlled protocols while others are opportunistic.
In this study[4], the authors use a collection of databases covering a range of species as well as of geographic scales in France and using different data collection and validation approaches as a case study to evaluate the impact of data quality when performing Strategic Environmental Assessment (SEA). Among their conclusions, the fact that a large-scale database (what they call the “country” level) is necessary to reliably parameterize SDM. Besides this and other conclusions of their study, which are likely to be in part specific to their case study –unfortunately for its conservation, biodiversity is complex and varies a lot–, the merit of this work lies in the approach used to test the impact of data on model predictions.
References
1. Feng, X. et al. A review of the heterogeneous landscape of biodiversity databases: Opportunities and challenges for a synthesized biodiversity knowledge base. Global Ecology and Biogeography 31, 1242–1260 (2022). https://doi.org/10.1111/geb.13497
2. Box, G. E. P. Robustness in the Strategy of Scientific Model Building. in Robustness in Statistics (eds. Launer, R. L. & Wilkinson, G. N.) 201–236 (Academic Press, 1979). https://doi.org/10.1016/B978-0-12-438150-6.50018-2.
3. Beissinger, S. R. & McCullough, D. R. Population Viability Analysis. (The University of Chicago Press, 2002).
4. Ferraille, T., Kerbiriou, C., Bigard, C., Claireau, F. & Thompson, J. D. (2023) Integrating biodiversity assessments into local conservation planning: the importance of assessing suitable data sources. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.05.09.539999