Direct submissions to PCI Ecology from bioRxiv.org are possible using the B2J service
Latest recommendations

| Id | Title * ▲ | Authors * | Abstract * | Picture * | Thematic fields * | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
14 Jan 2021
Consistent variations in personality traits and their potential for genetic improvement of biocontrol agents: Trichogramma evanescens as a case studySilène Lartigue, Myriam Yalaoui, Jean Belliard, Claire Caravel, Louise Jeandroz, Géraldine Groussier, Vincent Calcagno, Philippe Louâpre, François-Xavier Dechaume-Moncharmont, Thibaut Malausa and Jérôme Moreau https://doi.org/10.1101/2020.08.21.257881Tell us how you can be, and we’ll make you better: exploiting genetic variability in personality traits to improve top-down control of agricultural pestsRecommended by Marta Montserrat based on reviews by Bart A Pannebakker, François Dumont, Joshua Patrick Byrne and Ana Pimenta Goncalves PereiraAgriculture in the XXI century faces the huge challenge of having to provide food to a rapidly growing human population, which is expected to reach 10.9 billion in 2100 (UUNN 2019), by means of practices and methods that guarantee crop sustainability, human health safety, and respect to the environment (UUNN 2015). Such regulation by the United Nations ultimately entails that agricultural scientists are urged to design strategies and methods that effectively minimize the use of harmful chemical products to control pest populations and to improve soil quality. References Bielza, P., Balanza, V., Cifuentes, D. and Mendoza, J. E. (2020). Challenges facing arthropod biological control: Identifying traits for genetic improvement of predators in protected crops. Pest Manag Sci. doi: https://doi.org/10.1002/ps.5857 | Consistent variations in personality traits and their potential for genetic improvement of biocontrol agents: Trichogramma evanescens as a case study | Silène Lartigue, Myriam Yalaoui, Jean Belliard, Claire Caravel, Louise Jeandroz, Géraldine Groussier, Vincent Calcagno, Philippe Louâpre, François-Xavier Dechaume-Moncharmont, Thibaut Malausa and Jérôme Moreau | <p>Improvements in the biological control of agricultural pests require improvements in the phenotyping methods used by practitioners to select efficient biological control agent (BCA) populations in industrial rearing or field conditions. Consist... | Agroecology, Behaviour & Ethology, Biological control, Evolutionary ecology, Life history | Marta Montserrat | 2020-08-24 10:40:03 | View | ||
24 May 2022
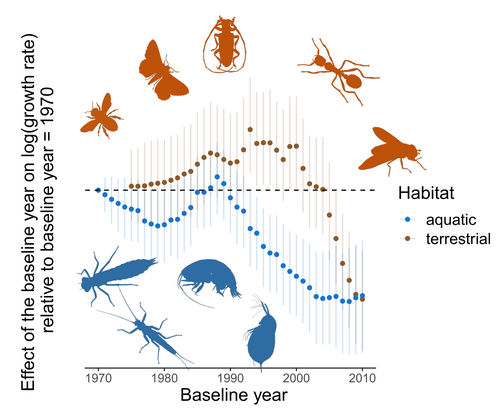
Controversy over the decline of arthropods: a matter of temporal baseline?François Duchenne, Emmanuelle Porcher, Jean-Baptiste Mihoub, Grégoire Loïs, Colin Fontaine https://doi.org/10.1101/2022.02.09.479422Don't jump to conclusions on arthropod abundance dynamics without appropriate dataRecommended by Tim Coulson based on reviews by Gabor L Lovei and 1 anonymous reviewer based on reviews by Gabor L Lovei and 1 anonymous reviewer
Humans are dramatically modifying many aspects of our planet via increasing concentrations of carbon dioxide in the atmosphere, patterns of land-use change, and unsustainable exploitation of the planet’s resources. These changes impact the abundance of species of wild organisms, with winners and losers. Identifying how different species and groups of species are influenced by anthropogenic activity in different biomes, continents, and habitats, has become a pressing scientific question with many publications reporting analyses of disparate data on species population sizes. Many conclusions are based on the linear analysis of rather short time series of organismal abundances. Duchenne F, Porcher E, Mihoub J-B, Loïs G, Fontaine C (2022) Controversy over the decline of arthropods: a matter of temporal baseline? bioRxiv, 2022.02.09.479422, ver. 3 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2022.02.09.479422 | Controversy over the decline of arthropods: a matter of temporal baseline? | François Duchenne, Emmanuelle Porcher, Jean-Baptiste Mihoub, Grégoire Loïs, Colin Fontaine | <p style="text-align: justify;">Recently, a number of studies have reported somewhat contradictory patterns of temporal trends in arthropod abundance, from decline to increase. Arthropods often exhibit non-monotonous variation in abundance over ti... |  | Conservation biology | Tim Coulson | 2022-02-11 15:44:44 | View | |
05 Nov 2019
Crown defoliation decreases reproduction and wood growth in a marginal European beech population.Sylvie Oddou-Muratorio, Cathleen Petit-Cailleux, Valentin Journé, Matthieu Lingrand, Jean-André Magdalou, Christophe Hurson, Joseph Garrigue, Hendrik Davi, Elodie Magnanou. https://doi.org/10.1101/474874Defoliation induces a trade-off between reproduction and growth in a southern population of BeechRecommended by Georges Kunstler based on reviews by 3 anonymous reviewersIndividuals ability to withstand abiotic and biotic stresses is crucial to the maintenance of populations at climate edge of tree species distribution. We start to have a detailed understanding of tree growth response and to a lesser extent mortality response in these populations. In contrast, our understanding of the response of tree fecundity and recruitment remains limited because of the difficulty to monitor it at the individual tree level in the field. Tree recruitment limitation is, however, crucial for tree population dynamics [1-2]. References [1] Clark, J. S. et al. (1999). Interpreting recruitment limitation in forests. American Journal of Botany, 86(1), 1-16. doi: 10.2307/2656950 | Crown defoliation decreases reproduction and wood growth in a marginal European beech population. | Sylvie Oddou-Muratorio, Cathleen Petit-Cailleux, Valentin Journé, Matthieu Lingrand, Jean-André Magdalou, Christophe Hurson, Joseph Garrigue, Hendrik Davi, Elodie Magnanou. | <p>1. Although droughts and heatwaves have been associated to increased crown defoliation, decreased growth and a higher risk of mortality in many forest tree species, their impact on tree reproduction and forest regeneration still remains underst... |  | Climate change, Eco-evolutionary dynamics, Molecular ecology, Physiology, Population ecology | Georges Kunstler | 2018-11-20 13:29:42 | View | |
01 Jun 2018
Data-based, synthesis-driven: setting the agenda for computational ecologyTimothée Poisot, Richard Labrie, Erin Larson, Anastasia Rahlin 10.1101/150128Some thoughts on computational ecology from people who I’m sure use different passwords for each of their accountsRecommended by Phillip P.A. Staniczenko based on reviews by Matthieu Barbier and 1 anonymous reviewerAre you an ecologist who uses a computer or know someone that does? Even if your research doesn’t rely heavily on advanced computational techniques, it likely hasn’t escaped your attention that computers are increasingly being used to analyse field data and make predictions about the consequences of environmental change. So before artificial intelligence and robots take over from scientists, now is great time to read about how experts think computers could make your life easier and lead to innovations in ecological research. In “Data-based, synthesis-driven: setting the agenda for computational ecology”, Poisot and colleagues [1] provide a brief history of computational ecology and offer their thoughts on how computational thinking can help to bridge different types of ecological knowledge. In this wide-ranging article, the authors share practical strategies for realising three main goals: (i) tighter integration of data and models to make predictions that motivate action by practitioners and policy-makers; (ii) closer interaction between data-collectors and data-users; and (iii) enthusiasm and aptitude for computational techniques in future generations of ecologists. The key, Poisot and colleagues argue, is for ecologists to “engage in meaningful dialogue across disciplines, and recognize the currencies of their collaborations.” Yes, this is easier said than done. However, the journey is much easier with a guide and when everyone involved serves to benefit not only from the eventual outcome, but also the process. References [1] Poisot, T., Labrie, R., Larson, E., & Rahlin, A. (2018). Data-based, synthesis-driven: setting the agenda for computational ecology. BioRxiv, 150128, ver. 4 recommended and peer-reviewed by PCI Ecology. doi: 10.1101/150128 | Data-based, synthesis-driven: setting the agenda for computational ecology | Timothée Poisot, Richard Labrie, Erin Larson, Anastasia Rahlin | Computational ecology, defined as the application of computational thinking to ecological problems, has the potential to transform the way ecologists think about the integration of data and models. As the practice is gaining prominence as a way to... |  | Meta-analyses, Statistical ecology, Theoretical ecology | Phillip P.A. Staniczenko | 2018-02-05 20:51:41 | View | |
04 Apr 2023
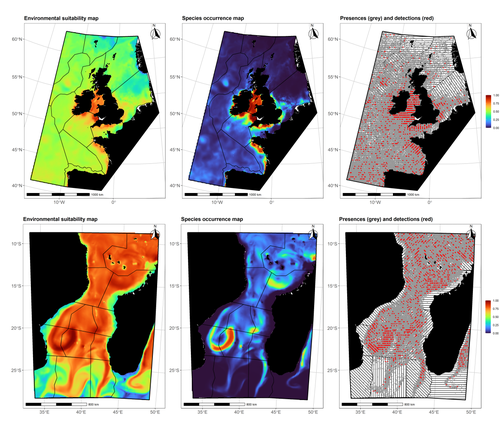
Data stochasticity and model parametrisation impact the performance of species distribution models: insights from a simulation studyCharlotte Lambert, Auriane Virgili https://doi.org/10.1101/2023.01.17.524386Species Distribution Models: the delicate balance between signal and noiseRecommended by Timothée Poisot based on reviews by Alejandra Zarzo Arias and 1 anonymous reviewer based on reviews by Alejandra Zarzo Arias and 1 anonymous reviewer
Species Distribution Models (SDMs) are one of the most commonly used tools to predict where species are, where they may be in the future, and, at times, what are the variables driving this prediction. As such, applying an SDM to a dataset is akin to making a bet: that the known occurrence data are informative, that the resolution of predictors is adequate vis-à-vis the scale at which their impact is expressed, and that the model will adequately capture the shape of the relationships between predictors and predicted occurrence. In this contribution, Lambert & Virgili (2023) perform a comprehensive assessment of different sources of complications to this process, using replicated simulations of two synthetic species. Their experimental process is interesting, in that both the data generation and the data analysis stick very close to what would happen in "real life". The use of synthetic species is particularly relevant to the assessment of SDM robustness, as they enable the design of species for which the shape of the relationship is given: in short, we know what the model should capture, and can evaluate the model performance against a ground truth that lacks uncertainty. Any simulation study is limited by the assumptions established by the investigators; when it comes to spatial data, the "shape" of the landscape, both in terms of auto-correlation and in where the predictors are available. Lambert & Virgili (2023) nicely circumvent these issues by simulating synthetic species against the empirical distribution of predictors; in other words, the species are synthetic, but the environment for which the prediction is made is real. This is an important step forward when compared to the use of e.g. neutral landscapes (With 1997), which can have statistical properties that are not representative of natural landscapes (see e.g. Halley et al., 2004). A striking point in the study by Lambert & Virgili (2023) is that they reveal a deep, indeed deeper than expected, stochasticity in SDMs; whether this is true in all models remains an open question, but does not invalidate their recommendation to the community: the interpretation of outcomes is a delicate exercise, especially because measures that inform on the goodness of the model fit do not capture the predictive quality of the model outputs. This preprint is both a call to more caution, and a call to more curiosity about the complex behavior of SDMs, while also providing a sensible template to perform future analyses of the potential issues with predictive models.
Halley, J. M., et al. (2004) “Uses and Abuses of Fractal Methodology in Ecology: Fractal Methodology in Ecology.” Ecology Letters, vol. 7, no. 3, pp. 254–71. https://doi.org/10.1111/j.1461-0248.2004.00568.x. Lambert, Charlotte, and Auriane Virgili (2023). Data Stochasticity and Model Parametrisation Impact the Performance of Species Distribution Models: Insights from a Simulation Study. bioRxiv, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.01.17.524386 With, Kimberly A. (1997) “The Application of Neutral Landscape Models in Conservation Biology. Aplicacion de Modelos de Paisaje Neutros En La Biologia de La Conservacion.” Conservation Biology, vol. 11, no. 5, pp. 1069–80. https://doi.org/10.1046/j.1523-1739.1997.96210.x. | Data stochasticity and model parametrisation impact the performance of species distribution models: insights from a simulation study | Charlotte Lambert, Auriane Virgili | <p>Species distribution models (SDM) are widely used to describe and explain how species relate to their environment, and predict their spatial distributions. As such, they are the cornerstone of most of spatial planning efforts worldwide. SDM can... |  | Biogeography, Habitat selection, Macroecology, Marine ecology, Spatial ecology, Metacommunities & Metapopulations, Species distributions, Statistical ecology | Timothée Poisot | 2023-01-20 09:43:51 | View | |
07 Oct 2019
Deer slow down litter decomposition by reducing litter quality in a temperate forestSimon Chollet, Morgane Maillard, Juliane Schorghuber, Sue Grayston, Jean-Louis Martin https://doi.org/10.1101/690032Disentangling effects of large herbivores on litter decompositionRecommended by Sébastien Barot based on reviews by 2 anonymous reviewersAboveground – belowground interactions is a fascinating field that has developed in ecology since about 20 years [1]. This field has been very fruitful as measured by the numerous articles published but also by the particular role it has played in the development of soil ecology. While soil ecology has for a long time developed partially independently from “general ecology” [2], the field of aboveground – belowground interactions has shown that all ecological interactions occurring within the soil are likely to impact plant growth and plant physiology because they have their roots within the soil. In turns, this should impact the aerial system of plants (higher or lower biomasses, changes in leaf quality…), which should cascade on the aboveground food web. Conversely, all ecological interactions occurring aboveground likely impact plant growth, which should cascade to their root systems, and thus to the soil functioning and the soil food web (through changes in the emission of exudates or inputs of dead roots…). Basically, plants are linking the belowground and aboveground worlds because, as terrestrial primary producers, they need to have (1) leaves to capture CO2 and exploit light and (2) roots to absorb water and mineral nutrients. The article I presently recommend [3] tackles this general issue through the prism of the impact of large herbivores on the decomposition of leaf litter. References [1] Hooper, D. U., Bignell, D. E., Brown, V. K., Brussard, L., Dangerfield, J. M., Wall, D. H. and Wolters, V. (2000). Interactions between Aboveground and Belowground Biodiversity in Terrestrial Ecosystems: Patterns, Mechanisms, and Feedbacks. BioScience, 50(12), 1049-1061. doi: 10.1641/0006-3568(2000)050[1049:ibaabb]2.0.co;2 | Deer slow down litter decomposition by reducing litter quality in a temperate forest | Simon Chollet, Morgane Maillard, Juliane Schorghuber, Sue Grayston, Jean-Louis Martin | <p>In temperate forest ecosystems, the role of deer in litter decomposition, a key nutrient cycling process, remains debated. Deer may modify the decomposition process by affecting plant cover and thus modifying litter abundance. They can also alt... | Community ecology, Ecosystem functioning, Herbivory, Soil ecology | Sébastien Barot | 2019-07-04 14:30:19 | View | ||
28 Dec 2022

Deleterious effects of thermal and water stresses on life history and physiology: a case study on woodlouseCharlotte Depeux, Angele Branger, Theo Moulignier, Jérôme Moreau, Jean-Francois Lemaitre, Francois-Xavier Dechaume-Moncharmont, Tiffany Laverre, Hélène Paulhac, Jean-Michel Gaillard, Sophie Beltran-Bech https://doi.org/10.1101/2022.09.26.509512An experimental approach for understanding how terrestrial isopods respond to environmental stressorsRecommended by Aniruddha Belsare based on reviews by Aaron Yilmaz and Michael Morris based on reviews by Aaron Yilmaz and Michael Morris
In this article, the authors discuss the results of their study investigating the effects of heat stress and moisture stress on a terrestrial isopod Armadilldium vulgare, the common woodlouse [1]. Specifically, the authors have assessed how increased temperature or decreased moisture affects life history traits (such as growth, survival, and reproduction) as well as physiological traits (immune cell parameters and \( beta \)-galactosidase activity). This article quantitatively evaluates the effects of the two stressors on woodlouse. Terrestrial isopods like woodlouse are sensitive to thermal and moisture stress [2; 3] and are therefore good models to test hypotheses in global change biology and for monitoring ecosystem health. An important feature of this study is the combination of experimental, laboratory, and analytical techniques. Experiments were conducted under controlled conditions in the laboratory by modulating temperature and moisture, life history and physiological traits were measured/analyzed and then tested using models. Both stressors had negative impacts on survival and reproduction of woodlouse, and result in premature ageing. Although thermal stress did not affect survival, it slowed woodlouse growth. Moisture stress did not have a detectable effect on woodlouse growth but decreased survival and reproductive success. An important insight from this study is that effects of heat and moisture stressors on woodlouse are not necessarily linear, and experimental approaches can be used to better elucidate the mechanisms and understand how these organisms respond to environmental stress. This article is timely given the increasing attention on biological monitoring and ecosystem health. References: [1] Depeux C, Branger A, Moulignier T, Moreau J, Lemaître J-F, Dechaume-Moncharmont F-X, Laverre T, Pauhlac H, Gaillard J-M, Beltran-Bech S (2022) Deleterious effects of thermal and water stresses on life history and physiology: a case study on woodlouse. bioRxiv, 2022.09.26.509512., ver. 3 peer-reviewd and recommended by PCI Ecology. https://doi.org/10.1101/2022.09.26.509512 [2] Warburg MR, Linsenmair KE, Bercovitz K (1984) The effect of climate on the distribution and abundance of isopods. In: Sutton SL, Holdich DM, editors. The Biology of Terrestrial Isopods. Oxford: Clarendon Press. pp. 339–367. [3] Hassall M, Helden A, Goldson A, Grant A (2005) Ecotypic differentiation and phenotypic plasticity in reproductive traits of Armadillidium vulgare (Isopoda: Oniscidea). Oecologia 143: 51–60. https://doi.org/10.1007/s00442-004-1772-3 | Deleterious effects of thermal and water stresses on life history and physiology: a case study on woodlouse | Charlotte Depeux, Angele Branger, Theo Moulignier, Jérôme Moreau, Jean-Francois Lemaitre, Francois-Xavier Dechaume-Moncharmont, Tiffany Laverre, Hélène Paulhac, Jean-Michel Gaillard, Sophie Beltran-Bech | <p>We tested independently the influences of increasing temperature and decreasing moisture on life history and physiological traits in the arthropod <em>Armadillidium vulgare</em>. Both increasing temperature and decreasing moisture led individua... |  | Biodiversity, Evolutionary ecology, Experimental ecology, Life history, Physiology, Terrestrial ecology, Zoology | Aniruddha Belsare | 2022-09-28 13:13:47 | View | |
10 Oct 2018

Detecting within-host interactions using genotype combination prevalence dataSamuel Alizon, Carmen Lía Murall, Emma Saulnier, Mircea T Sofonea https://doi.org/10.1101/256586Combining epidemiological models with statistical inference can detect parasite interactionsRecommended by Dustin Brisson based on reviews by Samuel Díaz Muñoz, Erick Gagne and 1 anonymous reviewerThere are several important topics in the study of infectious diseases that have not been well explored due to technical difficulties. One such topic is pursued by Alizon et al. in “Modelling coinfections to detect within-host interactions from genotype combination prevalences” [1]. Both theory and several important examples have demonstrated that interactions among co-infecting strains can have outsized impacts on disease outcomes, transmission dynamics, and epidemiology. Unfortunately, empirical data on pathogen interactions and their outcomes is often correlational making results difficult to decipher. References [1] Alizon, S., Murall, C.L., Saulnier, E., & Sofonea, M.T. (2018). Detecting within-host interactions using genotype combination prevalence data. bioRxiv, 256586, ver. 3 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/256586 | Detecting within-host interactions using genotype combination prevalence data | Samuel Alizon, Carmen Lía Murall, Emma Saulnier, Mircea T Sofonea | <p>Parasite genetic diversity can provide information on disease transmission dynamics but most methods ignore the exact combinations of genotypes in infections. We introduce and validate a new method that combines explicit epidemiological modelli... |  | Eco-immunology & Immunity, Epidemiology, Host-parasite interactions, Statistical ecology | Dustin Brisson | Samuel Díaz Muñoz, Erick Gagne | 2018-02-01 09:23:26 | View |
03 Jan 2024
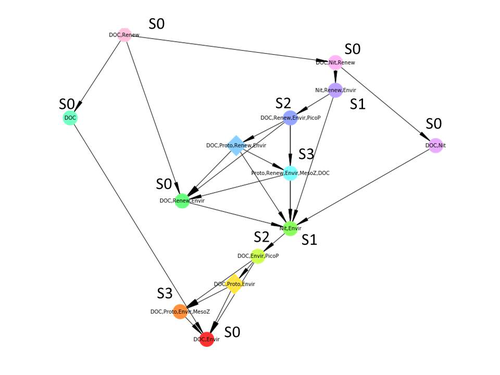
Diagnosis of planktonic trophic network dynamics with sharp qualitative changesCedric Gaucherel, Stolian Fayolle, Raphael Savelli, Olivier Philippine, Franck Pommereau, Christine Dupuy https://doi.org/10.1101/2023.06.29.547055A new approach to describe qualitative changes of complex trophic networksRecommended by Francis Raoul based on reviews by Tim Coulson and 1 anonymous reviewerModelling the temporal dynamics of trophic networks has been a key challenge for community ecologists for decades, especially when anthropogenic and natural forces drive changes in species composition, abundance, and interactions over time. So far, most modelling methods fail to incorporate the inherent complexity of such systems, and its variability, to adequately describe and predict temporal changes in the topology of trophic networks. Taking benefit from theoretical computer science advances, Gaucherel and colleagues (2024) propose a new methodological framework to tackle this challenge based on discrete-event Petri net methodology. To introduce the concept to naïve readers the authors provide a useful example using a simplistic predator-prey model. The core biological system of the article is a freshwater trophic network of western France in the Charente-Maritime marshes of the French Atlantic coast. A directed graph describing this system was constructed to incorporate different functional groups (phytoplankton, zooplankton, resources, microbes, and abiotic components of the environment) and their interactions. Rules and constraints were then defined to, respectively, represent physiochemical, biological, or ecological processes linking network components, and prevent the model from simulating unrealistic trajectories. Then the full range of possible trajectories of this mechanistic and qualitative model was computed. The model performed well enough to successfully predict a theoretical trajectory plus two trajectories of the trophic network observed in the field at two different stations, therefore validating the new methodology introduced here. The authors conclude their paper by presenting the power and drawbacks of such a new approach to qualitatively model trophic networks dynamics. Reference Cedric Gaucherel, Stolian Fayolle, Raphael Savelli, Olivier Philippine, Franck Pommereau, Christine Dupuy (2024) Diagnosis of planktonic trophic network dynamics with sharp qualitative changes. bioRxiv 2023.06.29.547055, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.06.29.547055 | Diagnosis of planktonic trophic network dynamics with sharp qualitative changes | Cedric Gaucherel, Stolian Fayolle, Raphael Savelli, Olivier Philippine, Franck Pommereau, Christine Dupuy | <p>Trophic interaction networks are notoriously difficult to understand and to diagnose (i.e., to identify contrasted network functioning regimes). Such ecological networks have many direct and indirect connections between species, and these conne... |  | Community ecology, Ecosystem functioning, Food webs, Freshwater ecology, Interaction networks, Microbial ecology & microbiology | Francis Raoul | Tim Coulson | 2023-07-03 10:42:34 | View |
30 Jan 2020
Diapause is not selected as a bet-hedging strategy in insects: a meta-analysis of reaction norm shapesJens Joschinski and Dries Bonte 10.1101/752881When to diapause or not to diapause? Winter predictability is not the answerRecommended by Bastien Castagneyrol based on reviews by Kévin Tougeron, Md Habibur Rahman Salman and 1 anonymous reviewerWinter is a harsh season for many organisms that have to cope with food shortage and potentially lethal temperatures. Many species have evolved avoidance strategies. Among them, diapause is a resistance stage many insects use to overwinter. For an insect, it is critical to avoid lethal winter temperatures and thus to initiate diapause before winter comes, while making the most of autumn suitable climatic conditions [1,2]. Several cues can be used to appreciate that winter is coming, including day length and temperature [3]. But climate changes, temperatures rise and become more variable from year to year, which imposes strong pressure upon insect phenology [4]. How can insects adapt to changes in the mean and variance of winter onset? References [1] Dyck, H. V., Bonte, D., Puls, R., Gotthard, K., and Maes, D. (2015). The lost generation hypothesis: could climate change drive ectotherms into a developmental trap? Oikos, 124(1), 54–61. doi: 10.1111/oik.02066 | Diapause is not selected as a bet-hedging strategy in insects: a meta-analysis of reaction norm shapes | Jens Joschinski and Dries Bonte | Many organisms escape from lethal climatological conditions by entering a resistant resting stage called diapause, and it is essential that this strategy remains optimally timed with seasonal change. Climate change therefore exerts selection press... | Maternal effects, Meta-analyses, Phenotypic plasticity, Terrestrial ecology | Bastien Castagneyrol | 2019-09-20 11:47:47 | View |
MANAGING BOARD
Julia Astegiano
Tim Coulson
Anna Eklof
Dominique Gravel
François Massol
Ben Phillips
Cyrille Violle










