
POISOT Timothée
- Biological Sciences, Universite de Montreal, Montreal, Canada
- Biodiversity, Biogeography, Climate change, Community ecology, Conservation biology, Ecosystem functioning, Epidemiology, Evolutionary ecology, Food webs, Host-parasite interactions, Interaction networks, Macroecology, Parasitology, Spatial ecology, Metacommunities & Metapopulations, Species distributions, Statistical ecology, Terrestrial ecology, Theoretical ecology
- recommender
Recommendations: 4
Reviews: 0
Recommendations: 4
Guidance framework to apply best practices in ecological data analysis: Lessons learned from building Galaxy-Ecology
Best practices for ecological analysis are required to act on concrete challenges
Recommended by Timothée Poisot based on reviews by Nick Isaac and 1 anonymous reviewerA core challenge facing ecologists is to work through an ever-increasing amount of data. The accelerating decline in biodiversity worldwide, mounting pressure of anthropogenic impacts, and increasing demand for actionable indicators to guide effective policy means that monitoring will only intensify, and rely on tools that can generate even more information (Gonzalez et al., 2023). How, then, do we handle this new volume and diversity of data?
This is the question Royaux et al. (2024) are tackling with their contribution. By introducing both a conceptual ("How should we think about our work?") and an operational ("Here is a tool to do our work with") framework, they establish a series of best practices for the analysis of ecological data.
It is easy to think about best practices in ecological data analysis in its most proximal form: is it good statistical practice? Is the experimental design correct? These have formed the basis of many recommendations over the years (see e.g. Popovic et al., 2024, for a recent example). But the contribution of Royaux et al. focuses on a different part of the analysis pipeline: the computer science (and software engineering) aspect of it.
As data grows in volume and complexity, the code needed to handle it follows the same trend. It is not a surprise, therefore, to see that the demand for programming skills in ecologists has doubled recently (Feng et al., 2020), prompting calls to make computational literacy a core component of undergraduate education (Farrell & Carrey, 2018). But beyond training, an obvious way to make computational analysis ecological data more reliable and effective is to build better tools. This is precisely what Royaux et al. have achieved.
They illustrate their approach through their experience building Galaxy-Ecology, a computing environment for ecological analysis: by introducing a clear taxonomy of computing concepts (data exploration, pre-processing, analysis, representation), with a hierarchy between them (formatting, data correction, anonymization), they show that we can think about the pipeline going from data to results in a way that is more systematized, and therefore more prone to generalization.
We may buckle at the idea of yet another ontology, or yet another framework, for our work, but I am convinced that the work of Royaux et al. is precisely what our field needs. Because their levels of atomization (their term for the splitting of complex pipelines into small, single-purpose tasks) are easy to understand, and map naturally onto tasks that we already perform, it is likely to see wide adoption. Solving the big, existential challenges of monitoring and managing biodiversity at the global scale requires the adoption of good practices, and a tool like Galaxy-Ecology goes a long way towards this goal.
References
Farrell, K.J., and Carey, C.C. (2018). Power, pitfalls, and potential for integrating computational literacy into undergraduate ecology courses. Ecol. Evol. 8, 7744-7751.
https://doi.org/10.1002/ece3.4363
Feng, X., Qiao, H., and Enquist, B. (2020). Doubling demands in programming skills call for ecoinformatics education. Frontiers in Ecology and the Environment 18, 123-124.
https://doi.org/10.1002/fee.2179
Gonzalez, A., Vihervaara, P., Balvanera, P., Bates, A.E., Bayraktarov, E., Bellingham, P.J., Bruder, A., Campbell, J., Catchen, M.D., Cavender-Bares, J., et al. (2023). A global biodiversity observing system to unite monitoring and guide action. Nat. Ecol. Evol., 1-5.
https://doi.org/10.1038/s41559-023-02171-0
Popovic, G., Mason, T.J., Drobniak, S.M., Marques, T.A., Potts, J., Joo, R., Altwegg, R., Burns, C.C.I., McCarthy, M.A., Johnston, A., et al. (2024). Four principles for improved statistical ecology. Methods Ecol. Evol. 15, 266-281.
https://doi.org/10.1111/2041-210X.14270
Coline Royaux, Jean-Baptiste Mihoub, Marie Jossé, Dominique Pelletier, Olivier Norvez, Yves Reecht, Anne Fouilloux, Helena Rasche, Saskia Hiltemann, Bérénice Batut, Marc Eléaume, Pauline Seguineau, Guillaume Massé, Alan Amossé, Claire Bissery, Romain Lorrilliere, Alexis Martin, Yves Bas, Thimothée Virgoulay, Valentin Chambon, Elie Arnaud, Elisa Michon, Clara Urfer, Eloïse Trigodet, Marie Delannoy, Gregoire Loïs, Romain Julliard, Björn Grüning, Yvan Le Bras (2024) Guidance framework to apply best practices in ecological data analysis: Lessons learned from building Galaxy-Ecology. EcoEvoRxiv, ver.3 peer-reviewed and recommended by PCI Ecology.
https://doi.org/10.32942/X2G033
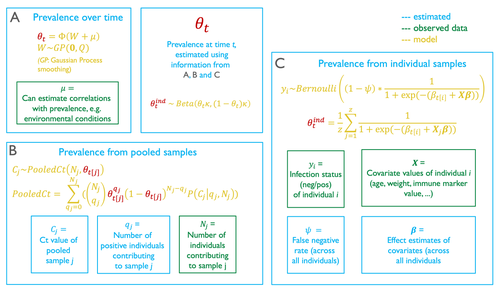
Reconstructing prevalence dynamics of wildlife pathogens from pooled and individual samples
Pooled samples hold information about the prevalence of wildlife pathogens
Recommended by Timothée Poisot based on reviews by Megan Griffiths and 2 anonymous reviewersAlthough monitoring the prevalence of pathogens in wildlife is crucial, there are logistical constraints that make this difficult, costly, and unpractical. This problem is often compounded when attempting to measure the temporal dynamics of prevalence. To improve the detection rate, a commonly used technique is pooling samples, where multiple individuals are analyzed at once. Yet, this introduces further potential biases: low-prevalence samples are effectively diluted through pooling, creating a false negative risk; negative samples are masked by the inclusion of positive samples, possibly artificially inflating the estimate of prevalence (and masking the inter-sample variability).
In their contribution, Borremans et al. (2024) come up with a modelling technique to provide accurate predictions of prevalence dynamics using a mix of pooled and individual samples. Because this model represents the pooling of individual samples as a complete mixing process, it can accurately estimate the prevalence dynamics from pooled samples only.
It is particularly noteworthy that the model provides an estimation of the false negative rate of the test. When there are false negatives (or more accurately, when the true rate at which false negatives happens), the value of the effect coefficients for individual-level covariates are likely to be off, potentially by a substantial amount. But besides more accurate coefficient estimation, the actual false negative rate is important information about the overall performance of the infection test.
The model described in this article also allows for a numerical calculation of the probability density function of infection. It is worth spending some time on how this is achieved, as I found the approach relying on combinatorics to be particularly interesting. When pooling, both the number of individuals that are mixed is known, and so is the measurement made on the pooled samples. The question is to figure out the number of individuals that because they are infectious, contribute to this score. The approach used by the authors is to draw (with replacement) possible positive and negative test outcomes assuming a number of positive individuals, and from this to estimate a pathogen concentration in the positive samples. This pathogen concentration can be transformed into its test outcome, and this value taken over all possible combinations is a conditional estimate of the test outcome, knowing the number of pooled individuals, and estimating the number of positive ones.
This approach is where the use of individual samples informs the model: by providing additional corrections for the relative volume of sample each individual provides, and by informing the transformation of test values into virus concentrations.
The authors make a strong case that their model can provide robust estimates of prevalence even in the presence of common field epidemiology pitfalls, and notably incomplete individual-level information. More importantly, because the model can work from pooled samples only, it gives additional value to samples that would otherwise have been discarded because they did not allow for prevalence estimates.
References
Benny Borremans, Caylee A. Falvo, Daniel E. Crowley, Andrew Hoegh, James O. Lloyd-Smith, Alison J. Peel, Olivier Restif, Manuel Ruiz-Aravena, Raina K. Plowright (2024) Reconstructing prevalence dynamics of wildlife pathogens from pooled and individual samples. bioRxiv, ver.3 peer-reviewed and recommended by PCI Ecology https://doi.org/10.1101/2023.11.02.565200
Modeling Tick Populations: An Ecological Test Case for Gradient Boosted Trees
Gradient Boosted Trees can deliver more than accurate ecological predictions
Recommended by Timothée Poisot based on reviews by 2 anonymous reviewersTick-borne diseases are an important burden on public health all over the globe, making accurate forecasts of tick population a key ingredient in a successful public health strategy. Over long time scales, tick populations can undergo complex dynamics, as they are sensitive to many non-linear effects due to the complex relationships between ticks and the relevant (numerical) features of their environment.
But luckily, capturing complex non-linear responses is a task that machine learning thrives on. In this contribution, Manley et al. (2023) explore the use of Gradient Boosted Trees to predict the distribution (presence/absence) and abundance of ticks across New York state.
This is an interesting modelling challenge in and of itself, as it looks at the same ecological question as an instance of a classification problem (presence/absence) or of a regression problem (abundance). In using the same family of algorithm for both, Manley et al. (2023) provide an interesting showcase of the versatility of these techniques. But their article goes one step further, by setting up a multi-class categorical model that estimates jointly the presence and abundance of a population. I found this part of the article particularly elegant, as it provides an intermediate modelling strategy, in between having two disconnected models for distribution and abundance, and having nested models where abundance is only predicted for the present class (see e.g. Boulangeat et al., 2012, for a great description of the later).
One thing that Manley et al. (2023) should be commended for is their focus on opening up the black box of machine learning techniques. I have never believed that ML models are more inherently opaque than other families of models, but the focus in this article on explainable machine learning shows how these models might, in fact, bring us closer to a phenomenological understanding of the mechanisms underpinning our observations.
There is also an interesting discussion in this article, on the rate of false negatives in the different models that are being benchmarked. Although model selection often comes down to optimizing the overall quality of the confusion matrix (for distribution models, anyway), depending on the type of information we seek to extract from the model, not all types of errors are created equal. If the purpose of the model is to guide actions to control vectors of human pathogens, a false negative (predicting that the vector is absent at a site where it is actually present) is a potentially more damaging outcome, as it can lead to the vector population (and therefore, potentially, transmission) increasing unchecked.
Boulangeat I, Gravel D, Thuiller W. Accounting for dispersal and biotic interactions to disentangle the drivers of species distributions and their abundances: The role of dispersal and biotic interactions in explaining species distributions and abundances. Ecol Lett. 2012;15: 584-593.
https://doi.org/10.1111/j.1461-0248.2012.01772.x
Manley W, Tran T, Prusinski M, Brisson D. (2023) Modeling tick populations: An ecological test case for gradient boosted trees. bioRxiv, 2023.03.13.532443, ver. 3 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.03.13.532443
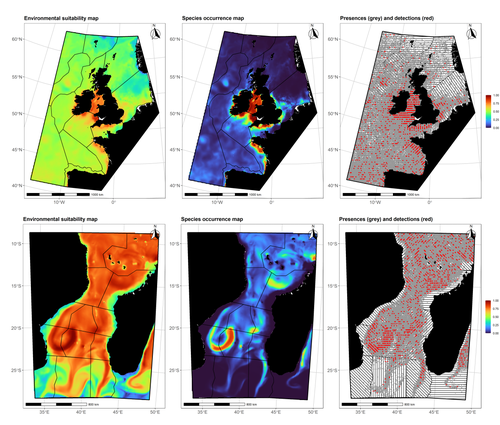
Data stochasticity and model parametrisation impact the performance of species distribution models: insights from a simulation study
Species Distribution Models: the delicate balance between signal and noise
Recommended by Timothée Poisot based on reviews by Alejandra Zarzo Arias and 1 anonymous reviewerSpecies Distribution Models (SDMs) are one of the most commonly used tools to predict where species are, where they may be in the future, and, at times, what are the variables driving this prediction. As such, applying an SDM to a dataset is akin to making a bet: that the known occurrence data are informative, that the resolution of predictors is adequate vis-à-vis the scale at which their impact is expressed, and that the model will adequately capture the shape of the relationships between predictors and predicted occurrence.
In this contribution, Lambert & Virgili (2023) perform a comprehensive assessment of different sources of complications to this process, using replicated simulations of two synthetic species. Their experimental process is interesting, in that both the data generation and the data analysis stick very close to what would happen in "real life". The use of synthetic species is particularly relevant to the assessment of SDM robustness, as they enable the design of species for which the shape of the relationship is given: in short, we know what the model should capture, and can evaluate the model performance against a ground truth that lacks uncertainty.
Any simulation study is limited by the assumptions established by the investigators; when it comes to spatial data, the "shape" of the landscape, both in terms of auto-correlation and in where the predictors are available. Lambert & Virgili (2023) nicely circumvent these issues by simulating synthetic species against the empirical distribution of predictors; in other words, the species are synthetic, but the environment for which the prediction is made is real. This is an important step forward when compared to the use of e.g. neutral landscapes (With 1997), which can have statistical properties that are not representative of natural landscapes (see e.g. Halley et al., 2004).
A striking point in the study by Lambert & Virgili (2023) is that they reveal a deep, indeed deeper than expected, stochasticity in SDMs; whether this is true in all models remains an open question, but does not invalidate their recommendation to the community: the interpretation of outcomes is a delicate exercise, especially because measures that inform on the goodness of the model fit do not capture the predictive quality of the model outputs. This preprint is both a call to more caution, and a call to more curiosity about the complex behavior of SDMs, while also providing a sensible template to perform future analyses of the potential issues with predictive models.
References
Halley, J. M., et al. (2004) “Uses and Abuses of Fractal Methodology in Ecology: Fractal Methodology in Ecology.” Ecology Letters, vol. 7, no. 3, pp. 254–71. https://doi.org/10.1111/j.1461-0248.2004.00568.x.
Lambert, Charlotte, and Auriane Virgili (2023). Data Stochasticity and Model Parametrisation Impact the Performance of Species Distribution Models: Insights from a Simulation Study. bioRxiv, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.01.17.524386
With, Kimberly A. (1997) “The Application of Neutral Landscape Models in Conservation Biology. Aplicacion de Modelos de Paisaje Neutros En La Biologia de La Conservacion.” Conservation Biology, vol. 11, no. 5, pp. 1069–80. https://doi.org/10.1046/j.1523-1739.1997.96210.x.