
A new approach to describe qualitative changes of complex trophic networks
Diagnosis of planktonic trophic network dynamics with sharp qualitative changes
Abstract
Recommendation: posted 02 January 2024, validated 03 January 2024
Raoul, F. (2024) A new approach to describe qualitative changes of complex trophic networks. Peer Community in Ecology, 100545. https://doi.org/10.24072/pci.ecology.100545
Recommendation
Modelling the temporal dynamics of trophic networks has been a key challenge for community ecologists for decades, especially when anthropogenic and natural forces drive changes in species composition, abundance, and interactions over time. So far, most modelling methods fail to incorporate the inherent complexity of such systems, and its variability, to adequately describe and predict temporal changes in the topology of trophic networks.
Taking benefit from theoretical computer science advances, Gaucherel and colleagues (2024) propose a new methodological framework to tackle this challenge based on discrete-event Petri net methodology. To introduce the concept to naïve readers the authors provide a useful example using a simplistic predator-prey model.
The core biological system of the article is a freshwater trophic network of western France in the Charente-Maritime marshes of the French Atlantic coast. A directed graph describing this system was constructed to incorporate different functional groups (phytoplankton, zooplankton, resources, microbes, and abiotic components of the environment) and their interactions. Rules and constraints were then defined to, respectively, represent physiochemical, biological, or ecological processes linking network components, and prevent the model from simulating unrealistic trajectories. Then the full range of possible trajectories of this mechanistic and qualitative model was computed.
The model performed well enough to successfully predict a theoretical trajectory plus two trajectories of the trophic network observed in the field at two different stations, therefore validating the new methodology introduced here. The authors conclude their paper by presenting the power and drawbacks of such a new approach to qualitatively model trophic networks dynamics.
Reference
Cedric Gaucherel, Stolian Fayolle, Raphael Savelli, Olivier Philippine, Franck Pommereau, Christine Dupuy (2024) Diagnosis of planktonic trophic network dynamics with sharp qualitative changes. bioRxiv 2023.06.29.547055, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.06.29.547055
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
SERVICESCALES project, from MetaProgram ECOSERV (INRAE, ECODIV department)
Reviewed by Tim Coulson , 30 Dec 2023
, 30 Dec 2023
My main criticism of the previous version of this manuscript is that it was hard going and difficult to follow. The careful rewrite, coupled with the alterations made in response to the specific comments of the other reviewer, now make this manuscript suitable to be recommended by PCI Ecology.
The method described is much clearer and appears to work in both theory and in practice. For this reason i believe the work provides a useful contribution to community ecology.
https://doi.org/10.24072/pci.ecology.100545.rev21Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/2023.06.29.547055
Version of the preprint: 1
Author's Reply, 05 Dec 2023
see attached files.
Decision by Francis Raoul, posted 02 Oct 2023, validated 03 Oct 2023
Dear authors,
Your manuscript has now been reviewed by two colleagues. They both point out that the method you propose could potentially be useful to the scientific community for modelling trophic networks. However, they both agree about strong flaws of the manuscript in its present form, to a point where it is quite hard to appreciate the added value of your work: (i) the modelling method and its validation process remain poorly explained and justified (vs. other options) and are not detailed enough for the work to be repeated by others, (ii) many terms are vague and/or left undefined so that it is hard to guess their meaning, (iii) the structure and flow of the article need to be revised, (iv) the English seems to need a strong editing.
I have decided to follow the reviewers’ opinion. I therefore encourage you to submit a new version of the manuscript that incorporates all suggestions made by them.
Best regards.
Reviewed by anonymous reviewer 1, 23 Aug 2023
Overall: The manuscript presents a fascinating topic, but the difficulty lies in introducing a new method to the field of ecology. This is an incredibly high challenge to meet, as it's more than "just" giving the Methods. The manuscript lacks structure, with a clear explanation of (1) the differences and advantages of the method pursued here, (2) a perfectly clear explanation of the method and walked-through application comparing to a well-known ecological example, (3) an explanation of what tests will be done to validate the method (how do we know it worked), and (4) guidelines given clearly in advance of what to expect for the Results section.
I cite 2 other papers below that manage to meet this challenge (De Cáceres et al. 2019; Silk et al. 2022), but there is a long line of new methods being introduced to ecologists then applied. I strongly suggest finding one such of these papers to use as a template, and to follow the structure used to guide what elements to include. I'm afraid as written, I can't truly evaluate the method and application. The manuscript read a bit more as a linear description - it was difficult to evaluate the model and its validation on data without knowing how the model worked and how the validation would be performed in advance of the Results section. Moreover, I simply don't find the Methods meets the standard of "Include sufficient details for the work to be repeated", which is critical for any study.
Abstract
Overall some typos (e.g. Line 29, "is assumed")
Line 19: Please be more precise about "to diagnose"
Line 20-21: Please be a bit more specific with "welcome numerous feedbacks" and "are not frozen". I appreciate the language choice, as I can infer what you may mean, but its much easier to be precise. For example. "Such ecological networks have many direct and indirect connections between species and these connections are not fixed, but often vary with time"
Line 23: dynamic (singular)
Line 24, 26: I can say again that the ambiguity of what "diagnosing" a network exactly means makes these two lines difficult - I am not sure yet what exactly will be done in the paper. (is it estimating species interaction strengths / connection strengths?)
Line 27: borrowing "from", not "to"
Line 32-34: I find this a bit speculative, and would prefer more quantitative conclusions. It should be a summary of the scientific findings
Introduction:
Lines 66: You write that equation-based models aren't well adapted to handle dynamical systems, but some theoretical CS tools "are able to handle" sharp changing networks - can you better summarise or articulate why one can't and another can? If you need to reduce space, I think the first 2 sentences in paragraph 2 could be deleted.
Line 67-69: Because you only introduced the field (theoretical CS) and the name of the approach (Petri net), the reader isn't brought in to understand exactly what you will do. Please better articulate the distinctions between the 2 approaches, a brief summary of what the theoretical CS (or Petri specifically) does. That will better set up the topic sentence you give here.
Line 107: Not clear what "well instrumented" means
Line 111: Same as above, the reader doesn't truly know what you will do given no explanation of the method and its distinction from existing approaches
Methods:
Overall - I'm afraid the delivery of these methods falls far short of what is needed to clearly understand how this type of model works. Many terms are left undefined, and even when they are defined, they are not always clear. A reader who sees "the rule is fired" might only make use of such a simplification if they already understand the entire process. I don't know exactly what edits to advise, but the section reads like someone explaining a very, very complicated figure and writing down what they say when explaining. Terms are only introduced right when they are critically needed to understand a given sentence, but no clear overall summary of how the methods works is given. The Petri net explanation could use much more structure, divided overall into the sections needed to understand this entire process.
It's not because this is complicated that it can't be clearly explained. See for example another paper for an Ecology audience that walks the reader through their method:
De Cáceres, M., Coll, L., Legendre, P., Allen, R. B., Wiser, S. K., Fortin, M.-J., Condit, R., and Hubbell, S.. 2019. Trajectory analysis in community ecology. Ecological Monographs 89(2):e01350. 10.1002/ecm.1350
I can only say again that I can't advise how to structure this, but it's quite difficult to follow how the method works. I would advise to share the Methods section with a group of PhD students, and ask for their feedback. Complicated methods can still be shared with ecology readers in a way that they can follow, but this explanation is too linear.
Line 115: For Figure 1 - (i) The figure is too difficult to read, please change for clarity. (ii) Please raise the position of the inset / left part of the figure - so that the total dimensions of the image become smaller. (iii) Pardon me if I am mistaken, but the figure appears to be associated with Google Maps - is it thus under some kind of copyright? Check that you have permissions needed to use such an image. However (iv) the image doesn't capture these are wetlands. Is there some kind of GIS layer that better shows the aquatic habitats? That information is more relevant than the road, town, and marine labels included. The text in lines 115-119 paint a rich aquatic picture of habitat and connectivity that the figure simply doesn't show.
Line 119-122: Trajectories of what? Also the description here doesn't inform the reader the date of the samples
Line 127: I didn't realise before this point that the trophic model would include environmental drivers - how can you better present this so we know to anticipate this? I argue above to give a better verbal preview of what the model is / does. Then here, you can simply list the information needed to reproduce the environmental measures. More detail needs to be given on how the model was constructed - where did this come from? What biological support is there for this model?
Line 127-141: I am not sure what to advise, but I really could not follow this verbal description of the model. It didn't help me understand Figure 2, and was difficult to read. I also advise that the information needed to follow the model (abbreviations etc) appear in the Figure 2 caption. In the Figure 2 caption, what does "upward" and "downward" mean? Also, you refer to abiotic components as "trophic"? This is very difficult for me to understand.
Line 150: What is "the leading question"?
Line 150-151: I find it awkward to be this far along and still not have any information on what a Petri net is.
Line 151: What does it mean to normalize a rule? I think there is a lack of description of what is done that is reproducible for a reader. This continues for Python automatically building a Petri net - how? Where are the full methods we need to evaluate this method, how does it build this?
I can see you give more detail in the subsequent sections, but it would be easier for you to state explicitly that this is a summary and more detail for each step is given subsequently.
Line 164: What is "2#nodes"?
Line 167: Its much too confusing to describe e.g. socioeconomic processes. We need the exact information needed to evaluate this study. I am struggling to really understand what methods this study uses, and having such general information at this point in the Methods is more confusing than clarifying.
Line 168-169: What does "neglectable" mean?
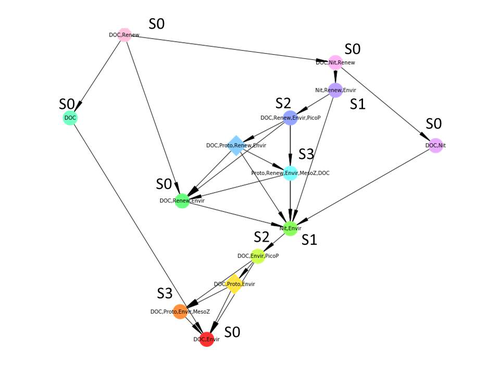
Lines 179: "were unlabelled arcs" - this sentence is not complete. Are the s0, s1, s2 all system states encountered? If so, indicate this abbreviation here in the legend. It's not clear to me why these are inactive rules. What is missing in Figure 3c between S0 and S1? It appears to be a "2" but the grey is partially blocked.
Line 182: What is a token circulation?
Line 186: What does "a realisation parts" mean?
Line 220: The reasoning for these 2 tests is not given.
Results:
Overall: I struggled (as I have indicated) that no general summary of the Petri net approach was given, before the detailed and difficult to follow explanation of the method. Many, many new terms are introduced and in some instances not defined. For this reason, it is almost impossible to know what to expect in the Results section. Sentences such as "when rule zero is deactivated, the model gets stuck into a high number of structural stabilities." I ask the authors to paint a better picture of what the model does, what the tests to the model consist of, and what the reader can expect to evaluate the results. It reads a bit too much like a descriptive report, and less like a systematic test of some hypothesis.
Line 261: How was this figure generated? Did the authors describe how they decide to merge state spaces? How does this figure help readers better understand trophic networks in plankton? I would suggest some kind of alternative, perhaps a Figure 1 that sketches a clear, well-known aquatic scenario, with some figure showing a traditional NPZ approach, and a 2nd figure showing a Petri approach, with some description of what can be learned using this alternate approach.
Line 271: This to me captures the difficulty I have with the manuscript - an "immature regime" is never defined. The amount of terms needed to follow grows too large, and I question what is gained by this approach. Even if you inform the reader an "immature regime" is a "biological winter", it means I search backwards until I can find what "biological winter" means. It is quite difficult to follow.
Line 293: I would again advise that the reader simply doesn't know what to expect when this very complex figure emerges. The method wasn't easy to follow, and only a very long list of Rules was given - what are the small black lines coming from the red nodes? How exactly can a plankton ecologist look at this figure and understand how it clarifies seasonal dynamics?
Line 301-302: It was confusing for me that this term was used in the Abstract, but only defined here.
Line 305-306: A difficulty is that the model is introduced to readers in a complex description, and proof isn't presented that the model can find states better than the experts identified here - how can we evaluate whether the trajectories identified by the model actually occurred?
Discussion:
Line 349-350: Because it is the 1st time to introduce this methods, the burden is on this manuscript to make absolutely sure the methods is perfectly clear to ecology (and not computer science) readers. There are other instances where new methods are introduced to ecologists, but special care must be given in this case. Here is one example:
https://onlinelibrary.wiley.com/doi/full/10.1111/ele.14079
Silk, M.J., Wilber, M.Q. & Fefferman, N.H. (2022) Capturing complex interactions in disease ecology with simplicial sets. Ecology Letters, 25, 2217–2231
https://doi.org/10.24072/pci.ecology.100545.rev11Reviewed by Tim Coulson , 20 Sep 2023
, 20 Sep 2023
The manuscript reports a new methodological approach based on directed graphs to parameterise temporally varying trophic networks. The approach is validated using a simulation, before it is then applied to real data. The approach does appear work well on the simple simulation, and does return results that appear sensible. Having said that, I did struggle to work out what was done.
The paper is hard going and rather poorly organised. I never like criticising language for researchers whose first language is not English, but this paper really needs review by a native English speaker. There are many places where I think translation from French has resulted in phrases that make little sense in English. As an example, the second line in the abstract states “Such ecological networks welcome numerous feedbacks between species and populations and are not frozen at all, as soon as we observe them over a long enough term.” The verb welcome is not correct, the frozen at all sentence refers to (I think) that most networks are treated as being static in time, but are dynamic. I could do this for most sentence in the manuscript. The clunky English make it very difficult to assess the work because it is hard to work out what has been done. I consequently found it rather hard to work out what has gone on.
If my understanding is correct, a directed graph of the trophic network is constructed. I can’t work out whether the initial graph contains all possible link, or is put together with expert knowledge. The initial model is then sampled using some sort of state-space formulation and a “best’ model identified. The model can contain temporal variation for each path in the model, and the temporal variation can be explained with an environmental driver. Is that correct?
I would be pleased to review a revised version of the manuscript once the English has been made more accessible. I think this will be important for the authors, because if the method is to become widely used, which is clearly the hope of the authors, the manuscript needs to be accessible. My summary is I think there is the glimmer of a great idea here, but it is very hard to assess and work out what has been done.
https://doi.org/10.24072/pci.ecology.100545.rev12









