
Some thoughts on computational ecology from people who I’m sure use different passwords for each of their accounts
Data-based, synthesis-driven: setting the agenda for computational ecology
Abstract
Recommendation: posted 01 June 2018, validated 01 June 2018
Staniczenko, P. (2018) Some thoughts on computational ecology from people who I’m sure use different passwords for each of their accounts. Peer Community in Ecology, 100002. https://doi.org/10.24072/pci.ecology.100002
Recommendation
Are you an ecologist who uses a computer or know someone that does? Even if your research doesn’t rely heavily on advanced computational techniques, it likely hasn’t escaped your attention that computers are increasingly being used to analyse field data and make predictions about the consequences of environmental change. So before artificial intelligence and robots take over from scientists, now is great time to read about how experts think computers could make your life easier and lead to innovations in ecological research. In “Data-based, synthesis-driven: setting the agenda for computational ecology”, Poisot and colleagues [1] provide a brief history of computational ecology and offer their thoughts on how computational thinking can help to bridge different types of ecological knowledge. In this wide-ranging article, the authors share practical strategies for realising three main goals: (i) tighter integration of data and models to make predictions that motivate action by practitioners and policy-makers; (ii) closer interaction between data-collectors and data-users; and (iii) enthusiasm and aptitude for computational techniques in future generations of ecologists. The key, Poisot and colleagues argue, is for ecologists to “engage in meaningful dialogue across disciplines, and recognize the currencies of their collaborations.” Yes, this is easier said than done. However, the journey is much easier with a guide and when everyone involved serves to benefit not only from the eventual outcome, but also the process.
References
[1] Poisot, T., Labrie, R., Larson, E., & Rahlin, A. (2018). Data-based, synthesis-driven: setting the agenda for computational ecology. BioRxiv, 150128, ver. 4 recommended and peer-reviewed by PCI Ecology. doi: 10.1101/150128
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
no declaration
Evaluation round #2
DOI or URL of the preprint: 10.1101/150128
Version of the preprint: 3
Author's Reply, 29 May 2018
Dear Editor,
Thank you for this in-depth reading of our manuscript. We have clarified all of the points, and fixed the language issues.
Best, Timothée Poisot
Decision by Phillip P.A. Staniczenko, posted 29 May 2018
Dear Timothée,
Thank you for sending a response to reviewer comments and a revised version of your preprint, "Data-based, synthesis-driven: setting the agenda for computational ecology."
I have read the revised preprint and would be happy to recommend the next version as part of PCI Ecology.
It is a very interesting thought piece about the role of computational ecology in ecological research, and I share your desire to encourage greater interaction between data-collectors and data-users. I hope this article will stimulate discussion among computational ecologists about the best way of advancing the field.
Below, I offer some thoughts of my own that you may want to consider as you move forward with the manuscript, along with some minor comments primarily with suggestions about spelling and grammar.
Please make some minor revisions and submit an updated version to PCI Ecology, at which point I will recommend the preprint and write a supporting statement.
Best wishes, Phillip
#
THOUGHTS
In the abstract, it might be nice to include a description of what you mean by "computational thinking."
It would helpful to include a paragraph near the end of the introduction that summarises your position and outlook for computational ecology (similar to what you have in the paragraph at the top of Page 8). This paragraph could cover how you think computational ecology can be useful in the coming years: What kind of role will it have? What kind of problems and systems will it be useful for?
Around the second paragraph on Page 2, it might be nice to explain or provide definitions for different types of model---e.g., mechanistic, process-based, statistical, mathematical---and how they fit into your argument about computational ecology and linking data and models (you could also discuss the relationship between ecological theory and ecological models).
On Page 2, second paragraph, you raise the following interesting idea: "we highlight the ways in which computational ecology differs from, and complements, ecological modelling." Maybe I missed it, but I didn't find where you addressed this point in the manuscript? By "ecological modelling" do you mean mathematical models that don't involve data?
On Page 2, third paragraph, you could make it clear that you are providing your viewpoint about computational ecology (or is the definition you provide in the opening sentence generally accepted in the field, in which case you should provide a reference).
On Page 4, third paragraph, when you say "recent examples" do you mean examples of successes, or examples that require improvement? Also, when you mention "RNA-seq data and worldclim climate data" are you referring to a single study that uses both of them together, or as two separate examples of something?
On Page 6, first paragraph, you talk about models and prediction. I hold the opinion that any mathematical (as distinct from conceptual) model can provide a prediction, including statistical models like GLMs. At this point in the text, there is an opportunity to discuss issues such as the relative importance of different models, how different models improve our understanding of the natural world, how well models describe or elucidate ecological process, and then how different models are actually used, i.e., the "so what" and "what next" questions.
On Page 7, first paragraph, you mention the paper by Thessen (2016)---it might be helpful to expand on your statement.
On Page 8, third paragraph, you could briefly define what you mean by "synthesis" in this context.
On Page 8, fourth paragraph, there is an opportunity to talk about how much accuracy is required of predictions for different purposes, e.g., science compared to policy and decision-making.
MINOR COMMENTS
Abstract. "broader landscape of ecological research."
Abstract. "one another's expertise"
Page 1. Personally, I don't love that definition of a complex problem, but you could include a reference in support of that statement.
Page 1. Regarding the definition of computational science: to me it seems to exclude machine learning---do you think that's the case?
Page 2. Very top: missing closing parenthesis after "completeness"
Page 2. Second paragraph: "reasons for why theoretical papers"
Page 2. Consider use of "deficit"---"experience relatively lower numbers of citations"?
Page 2. Second paragraph: "leading textbooks on mathematical models"
Page 2. Third paragraph: "stand on their own"
Page 3. First paragraph: "mechanisms of interest"
Page 3. Second paragraph: "one field has a high potential"
Page 3. Second paragraph: offer a definition of "synthesis" and why it is desirable.
Page 3. Third paragraph: "species distribution modelling"; "presences and absences"; "in which the observations were made"
Page 5. Second paragraph: "classic predator-prey equations"
Page 5. Fifth paragraph: "Predator-prey interactions"
Page 6. First paragraph: "to an extent"
Page 6. Second paragraph: "predator-prey interactions"
Page 9. Second paragraph: clarify that it is the uptake that is abysmal; "ultimately limited because"
Page 9. Third paragraph: "propagate form one analysis to another"; "in a succession of predictive steps"; explain what you mean by "de novo"
Page 10. Third paragraph: "scale of individual researchers"
Page 10. Fourth paragraph: "purpose they were produced for."
Page 10. Fifth paragraph: "different scales to address"
Page 11. Second paragraph: "needs these data"
Page 11. Third paragraph: "fruitful avenues"
Page 12. First paragraph: "does of course require discussion"
Page 12. Second paragraph: "higher standards"; just checking meaning: "programmatic approaches" does not exclusively mean approaches involving (computer) programming, rather, it means any approach that follows an established sequence of steps.
Page 12. Third paragraph: "alongside more reproducible publication"; "reproducible science"; "and show how data were transformed"
Page 12. Fifth paragraph: Which "approaches" are you referring to? Because you also say that "direct observation" and "experimentation" are both intrinsically superior (than other approaches).
Page 13. First paragraph: "under the conditions"
Page 13. Third paragraph: "allows the generation of predictions"
Finally, please check all references are complete and reasonably formatted.
Evaluation round #1
DOI or URL of the preprint: 10.1101/150128
Version of the preprint: 2
Author's Reply, 08 May 2018
Dear recommender,
We have completed a revision of our preprint. It should be updated on biorxiv shortly, but we have provided a copy here.
We re-wrote almost all of section 2, and re-designed all figures. This is an important change to the manuscript, but we believe it solves most of the points by reviewers.
We have also greatly expanded the scope of examples from the literature we give.
Decision by Phillip P.A. Staniczenko, posted 08 May 2018
Dear authors,
I have now received two reviews of your manuscript. Both reviewers find your manuscript interesting, but also raise issues that should be addressed. I suggest revising your manuscript in light of their comments, accompanied by a thorough response to their reviews.
I look forward to receiving a revised version of your manuscript.
Yours sincerely,
Phillip Staniczenko
#
Reviewer 1
The manuscript by Poisot et al. is an opinion/synthesis paper about “computational ecology”. The authors explain why it is crucial to develop this “new” field and describe how this should be achieved. In particular, the authors emphasize rightly the need to better link ecological data and computer models.
Thought I find this manuscript interesting I have three types of general criticism:
(1) Is all this very new? My understanding is that 20 years ago a large part of the scientific community working in ecology was somehow despising complex computer models, basically because these models were thought as too complicated to help understanding reality and because computer programs were deemed not enough reliable… However, I think that already for some years the community has been recognizing the value of computer models to increase ecological understanding and that some papers have already pointed out the value of these models.
(2) Many points are interesting in the manuscript but are not enough supported by detailed arguments or examples. For example, it is explained that mathematical models cannot take into account the complexity of ecological systems, e.g. stochasticity, but it is not fully explained why. Similarly, no successful example of computational ecology project is detailed enough to fully convince of the value of computational ecology. The authors also rise rightfully about important issues, e.g. the need to increase data sharing, but do not really give new solutions to these problems.
(3) I see all approaches in ecology as a continuum and the only way for me to develop ecology is to recognize the usefulness and shortcomings of all approaches and to use these approaches in a complementary way. I find that somehow the manuscript does not recognize enough that mathematical models in ecology have some virtue and that conversely computer models have some drawbacks. For example, theoretical mathematical model are hard to parameterize and their results are difficult to compare quantitatively to real data, but qualitative comparisons are also useful. Computer models are very flexible and very good at making precise quantitative predictions but these predictions require a huge amount of data for each new ecosystem / species for which predictions have to be made.
Reviewer 2
SUMMARY
Section 1 gives Species Distribution Models as a flagship example of computational ecology: having started as purely statistical, this field incorporates more and more theoretical concepts, and is implied (more than said, though that may qualify as common knowledge) to be rather heavily computational in the types of tools and practices employed to do so.
Section 2 attempts to better define the boundaries of computational ecology and its relationship to other branches of ecological research. Mainly, integration of data and models emerges as the determining factor; a second distinction of suggesting rather than documenting mechanisms is proposed, but its concrete implications are less clear.
Section 3 then lists recommendations toward ecological synthesis and collaboration, which largely fall into two bins:
data best practices, including metadata and common formats, reproducibility, data sharing and how to foster and credit it.
sensitivity training to quantitative skills, including fostering a healthy (and critical) relationship to code and software among empiricists, as well as more dialogue between the three poles of empirism, computation and theory.
More in-passing remarks concern the ability of computational ecology to provide tools relaying fundamental research toward its many applications (predictions for "action ecology" and stakeholders, etc) This paper starkly (sometimes wittily) states a number of problems, and emphasizes important directions in solving them (such as the problem of properly crediting and encouraging data producers, as their data increasingly must be reused beyond what they could hope to achieve on their own).
GENERAL THOUGHTS:
This is an enjoyable read, with a worthwhile discussion of the conditions under which computational endeavors in ecology can exist, prosper and expand the field in general. The calls for 1) more open, inter-communicable and reusable (hence far less wasteful in terms of scientific effort) data practices and 2) more integration of data and theory (in training as well as research), are both very important and ever timely.
Being myself computationally minded, and therefore part of the cheer squad here, I defer to more worldly ecologists on the question of whether this paper's content is adequate for its goals. I do wonder if these concerns have not been voiced before in a similar fashion, and whether this paper is constructed so as to change minds that were not already on the authors' side. Of course, it is always useful to restate the problems at stake here, and the review-like quality of parts of this paper provides a trove of references and semi-concrete suggestions.
Less importantly, I am not so clear on what the initial sections, trying to define or characterize computational ecology, really achieve, and in particular how computational ecology is meant to differ from any theoretical work that is not entirely data-free.
An obvious distinction would be a pragmatic one of techniques and training - crudely, a question of volume of data (and code required to handle said data) - rather than content. But I feel that the paper is also trying to suggest such a distinction of content, which I cannot quite grasp. For instance, would Hubbell's neutral theory, or Maximum Entropy style models, be considered computational ecology? If they do have such a distinction in mind, I would like the authors to find a starker way to express it, as it could improve the dialogue between computational and theoretical ecologists in the future (an interface which is far less addressed than the one with empiricists).
Some questions:
What's new since Pascual 2005? Many references and specific proposals listed here are recent, but has the field of computational ecology in general changed shape or direction somewhat since then?
Optionally, I think it would be interesting to provide some discussion of epidemiology here, since I feel it already does (for a certain type of population dynamics) a lot of what is proposed for the intersection of "action" and computational ecology, in particular short-cycle predictions, tools for stakeholders, and so on. Looking at these practices, if they are not too far removed from the authors' field of expertise, could help make new concrete suggestions, and perhaps recognize some opportunities or problems that computational ecology may face in the future.
MINOR CONTENT COMMENTS:
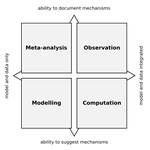
Fig 1: The locations (or labels) of meta-analysis and observation are a bit perplexing at first. If anything, I would expect the typical observational work to have typically less integration with models than a meta-analysis (depending on when statistics qualify as modelling in your perspective, which seems to me to oscillate a bit throughout the text) If by "observation" you mean the paragraph about Sallan et al., then I understand better, but I would suggest making the relationship between the figure and the main text more obvious. Perhaps due to this, the paragraph on meta-analyses in page 6 seems slightly disconnected from the rest.
Table 1: I am not sure what you mean by accuracy, for it to be high for mathematical models, observation and meta-analysis all at once. I am also unsure about what is "indirect" suitability for prediction. As for the invisible costs of computational infrastructure, I can see that they are immense at the social level, as noted at the end of the discussion, but its seems to me that most of these costs are not borne by researchers and therefore not really relevant to choosing a research practice (obviously pure math is somewhat cheaper, but to caricature, even a pure mathematician requires a lot of infrastructure to access publications from across the world, to say nothing of chalk and coffee)
page 8: I would like a bit more clarity on Ovaskainen's proposal (translating ecological concepts into digital representations); right now it seems very vague.
Paragraph "With or without a common data format..." I am not clear on what you have in mind here, talking about prediction propagating "from one analysis to the other" Do you mean the fact that computational ecology reuses pre-analyzed rather than raw data? That a single computational ecology project itself tends to have multiple analytical steps? Or somtehing else?
WRITING
Throughout, you probably mean "quadrant" rather than "quadrat", although this is a very ecological slip to make. Of course, if this is conscious and tongue-in-cheek, feel free to go with it.
page 2 Computational science is one of the ways to practice computational thinking: Makes me really perplex about what the other ways are "to the point where one would almost..." : That's a very mild statement "ecological systems as complex and adaptive [...] a certain degree of stochasticity" -> the link is not so obvious. "modelling, that can often function on {its} own"
page 3: P(S|E=1) should be P(S=1|E) de novo field samplig -> sampling
page 6: "it is paradoxically the high degree of model abstraction ... that gives them generality". It is not entirely clear to me how 1) this is obviously paradoxical, depending on hwat is meant by generality, and 2) this paradox is explained by the sentences that follow (they are about the fact that data is hard to collect, which is tangentially relevant).
page7 computaitonal research is dependant-> ent
page9: I simply wish to wholeheartedly support this beautiful defense of parasites. conservation target -> targetS
page 10: "and how data were transformed during the analysis" is probably lacking a verb before the "how".
Reviewed by Matthieu Barbier , 19 Feb 2018
, 19 Feb 2018
SUMMARY
Section 1 gives Species Distribution Models as a flagship example of computational ecology: having started as purely statistical, this field incorporates more and more theoretical concepts, and is implied (more than said, though that may qualify as common knowledge) to be rather heavily computational in the types of tools and practices employed to do so.
Section 2 attempts to better define the boundaries of computational ecology and its relationship to other branches of ecological research. Mainly, integration of data and models emerges as the determining factor; a second distinction of suggesting rather than documenting mechanisms is proposed, but its concrete implications are less clear.
Section 3 then lists recommendations toward ecological synthesis and collaboration, which largely fall into two bins: - data best practices, including metadata and common formats, reproducibility, data sharing and how to foster and credit it. - sensitivity training to quantitative skills, including fostering a healthy (and critical) relationship to code and software among empiricists, as well as more dialogue between the three poles of empirism, computation and theory. More in-passing remarks concern the ability of computational ecology to provide tools relaying fundamental research toward its many applications (predictions for "action ecology" and stakeholders, etc)
This paper starkly (sometimes wittily) states a number of problems, and emphasizes important directions in solving them (such as the problem of properly crediting and encouraging data producers, as their data increasingly must be reused beyond what they could hope to achieve on their own).
GENERAL THOUGHTS:
This is an enjoyable read, with a worthwhile discussion of the conditions under which computational endeavors in ecology can exist, prosper and expand the field in general. The calls for 1) more open, inter-communicable and reusable (hence far less wasteful in terms of scientific effort) data practices and 2) more integration of data and theory (in training as well as research), are both very important and ever timely.
Being myself computationally minded, and therefore part of the cheer squad here, I defer to more worldly ecologists on the question of whether this paper's content is adequate for its goals. I do wonder if these concerns have not been voiced before in a similar fashion, and whether this paper is constructed so as to change minds that were not already on the authors' side. Of course, it is always useful to restate the problems at stake here, and the review-like quality of parts of this paper provides a trove of references and semi-concrete suggestions.
Less importantly, I am not so clear on what the initial sections, trying to define or characterize computational ecology, really achieve, and in particular how computational ecology is meant to differ from any theoretical work that is not entirely data-free. An obvious distinction would be a pragmatic one of techniques and training - crudely, a question of volume of data (and code required to handle said data) - rather than content. But I feel that the paper is also trying to suggest such a distinction of content, which I cannot quite grasp. For instance, would Hubbell's neutral theory, or Maximum Entropy style models, be considered computational ecology? If they do have such a distinction in mind, I would like the authors to find a starker way to express it, as it could improve the dialogue between computational and theoretical ecologists in the future (an interface which is far less addressed than the one with empiricists).
Some questions:
What's new since Pascual 2005? Many references and specific proposals listed here are recent, but has the field of computational ecology in general changed shape or direction somewhat since then?
Optionally, I think it would be interesting to provide some discussion of epidemiology here, since I feel it already does (for a certain type of population dynamics) a lot of what is proposed for the intersection of "action" and computational ecology, in particular short-cycle predictions, tools for stakeholders, and so on. Looking at these practices, if they are not too far removed from the authors' field of expertise, could help make new concrete suggestions, and perhaps recognize some opportunities or problems that computational ecology may face in the future.
MINOR CONTENT COMMENTS:
Fig 1: The locations (or labels) of meta-analysis and observation are a bit perplexing at first. If anything, I would expect the typical observational work to have typically less integration with models than a meta-analysis (depending on when statistics qualify as modelling in your perspective, which seems to me to oscillate a bit throughout the text)
If by "observation" you mean the paragraph about Sallan et al., then I understand better, but I would suggest making the relationship between the figure and the main text more obvious. Perhaps due to this, the paragraph on meta-analyses in page 6 seems slightly disconnected from the rest.
Table 1: I am not sure what you mean by accuracy, for it to be high for mathematical models, observation and meta-analysis all at once. I am also unsure about what is "indirect" suitability for prediction.
As for the invisible costs of computational infrastructure, I can see that they are immense at the social level, as noted at the end of the discussion, but its seems to me that most of these costs are not borne by researchers and therefore not really relevant to choosing a research practice (obviously pure math is somewhat cheaper, but to caricature, even a pure mathematician requires a lot of infrastructure to access publications from across the world, to say nothing of chalk and coffee)
page 8: I would like a bit more clarity on Ovaskainen's proposal (translating ecological concepts into digital representations); right now it seems very vague.
Paragraph "With or without a common data format..." I am not clear on what you have in mind here, talking about prediction propagating "from one analysis to the other" Do you mean the fact that computational ecology reuses pre-analyzed rather than raw data? That a single computational ecology project itself tends to have multiple analytical steps? Or somtehing else?
WRITING
Throughout, you probably mean "quadrant" rather than "quadrat", although this is a very ecological slip to make. Of course, if this is conscious and tongue-in-cheek, feel free to go with it.
page 2 Computational science is one of the ways to practice computational thinking: Makes me really perplex about what the other ways are
"to the point where one would almost..." : That's a very mild statement
"ecological systems as complex and adaptive [...] a certain degree of stochasticity" -> the link is not so obvious.
"modelling, that can often function on {its} own"
page 3: P(S|E=1) should be P(S=1|E) de novo field samplig -> sampling
page 6: "it is paradoxically the high degree of model abstraction ... that gives them generality". It is not entirely clear to me how 1) this is obviously paradoxical, depending on hwat is meant by generality, and 2) this paradox is explained by the sentences that follow (they are about the fact that data is hard to collect, which is tangentially relevant).
page7 computaitonal research is dependant-> ent
page9: I simply wish to wholeheartedly support this beautiful defense of parasites.
conservation target -> targetS
page 10: "and how data were transformed during the analysis" is probably lacking a verb before the "how".
https://doi.org/10.24072/pci.ecology.100002.rev11Reviewed by anonymous reviewer 1, 12 Feb 2018
The manuscript by Poisot et al. is an opinion/synthesis paper about “computational ecology”. The authors explain why it is crucial to develop this “new” field and describe how this should be achieved. In particular, the authors emphasize rightly the need to better link ecological data and computer models. Thought I find this manuscript interesting I have three types of general criticism: (1) Is all this very new? My understanding is that 20 years ago a large part of the scientific community working in ecology was somehow despising complex computer models, basically because these models were thought as too complicated to help understanding reality and because computer programs were deemed not enough reliable… However, I think that already for some years the community has been recognizing the value of computer models to increase ecological understanding and that some papers have already pointed out the value of these models. (2) Many points are interesting in the manuscript but are not enough supported by detailed arguments or examples. For example, it is explained that mathematical models cannot take into account the complexity of ecological systems, e.g. stochasticity, but it is not fully explained why. Similarly, no successful example of computational ecology project is detailed enough to fully convince of the value of computational ecology. The authors also rise rightfully about important issues, e.g. the need to increase data sharing, but do not really give new solutions to these problems. (3) I see all approaches in ecology as a continuum and the only way for me to develop ecology is to recognize the usefulness and shortcomings of all approaches and to use these approaches in a complementary way. I find that somehow the manuscript does not recognize enough that mathematical models in ecology have some virtue and that conversely computer models have some drawbacks. For example, theoretical mathematical model are hard to parameterize and their results are difficult to compare quantitatively to real data, but qualitative comparisons are also useful. Computer models are very flexible and very good at making precise quantitative predictions but these predictions require a huge amount of data for each new ecosystem / species for which predictions have to be made.
https://doi.org/10.24072/pci.ecology.100002.rev12









