Direct submissions to PCI Ecology from bioRxiv.org are possible using the B2J service
Latest recommendations

| Id | Title * | Authors * | Abstract * | Picture * | Thematic fields * ▲ | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
18 Dec 2020

Once upon a time in the far south: Influence of local drivers and functional traits on plant invasion in the harsh sub-Antarctic islandsManuele Bazzichetto, François Massol, Marta Carboni, Jonathan Lenoir, Jonas Johan Lembrechts, Rémi Joly, David Renault https://doi.org/10.1101/2020.07.19.210880A meaningful application of species distribution models and functional traits to understand invasion dynamicsRecommended by Joaquín Hortal based on reviews by Paula Matos and Peter Convey based on reviews by Paula Matos and Peter Convey
Polar and subpolar regions are fragile environments, where the introduction of alien species may completely change ecosystem dynamics if the alien species become keystone species (e.g. Croll, 2005). The increasing number of human visits, together with climate change, are favouring the introduction and settling of new invaders to these regions, particularly in Antarctica (Hughes et al. 2015). Within this context, the joint use of Species Distribution Models (SDM) –to assess the areas potentially suitable for the aliens– with other measures of the potential to become successful invaders can inform on the need for devoting specific efforts to eradicate these new species before they become naturalized (e.g. Pertierra et al. 2016). References Austin, M. P., Nicholls, A. O., and Margules, C. R. (1990). Measurement of the realized qualitative niche: environmental niches of five Eucalyptus species. Ecological Monographs, 60(2), 161-177. doi: https://doi.org/10.2307/1943043 | Once upon a time in the far south: Influence of local drivers and functional traits on plant invasion in the harsh sub-Antarctic islands | Manuele Bazzichetto, François Massol, Marta Carboni, Jonathan Lenoir, Jonas Johan Lembrechts, Rémi Joly, David Renault | <p>Aim Here, we aim to: (i) investigate the local effect of environmental and human-related factors on alien plant invasion in sub-Antarctic islands; (ii) explore the relationship between alien species features and their dependence on anthropogeni... |  | Biogeography, Biological invasions, Spatial ecology, Metacommunities & Metapopulations, Species distributions | Joaquín Hortal | 2020-07-21 21:13:08 | View | |
13 May 2024
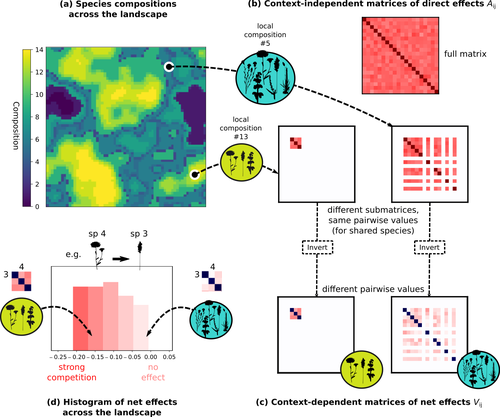
Getting More by Asking for Less: Linking Species Interactions to Species Co-Distributions in MetacommunitiesMatthieu Barbier, Guy Bunin, Mathew A. Leibold https://doi.org/10.1101/2023.06.04.543606Beyond pairwise species interactions: coarser inference of their joined effects is more relevantRecommended by François Munoz based on reviews by Frederik De Laender, Hao Ran Lai and Malyon Bimler based on reviews by Frederik De Laender, Hao Ran Lai and Malyon Bimler
Barbier et al. (2024) investigated the dynamics of species abundances depending on their ecological niche (abiotic component) and on (numerous) competitive interactions. In line with previous evidence and expectations (Barbier et al. 2018), the authors show that it is possible to robustly infer the mean and variance of interaction coefficients from species co-distributions, while it is not possible to infer the individual coefficient values. The authors devised a simulation framework representing multispecies dynamics in an heterogeneous environmental context (2D grid landscape). They used a Lotka-Volterra framework involving pairwise interaction coefficients and species-specific carrying capacities. These capacities depend on how well the species niche matches the local environmental conditions, through a Gaussian function of the distance of the species niche centers to the local environmental values. They considered two contrasted scenarios denoted as « Environmental tracking » and « Dispersal limited ». In the latter case, species are initially seeded over the environmental grid and cannot disperse to other cells, while in the former case they can disperse and possibly be more performant in other cells. The direct effects of species on one another are encoded in an interaction matrix A, and the authors further considered net interactions depending on the inverse of the matrix of direct interactions (Zelnik et al., 2024). The net effects are context-dependent, i.e., it involves the environment-dependent biotic capacities, even through the interaction terms can be defined between species as independent from local environment. The results presented here underline that the outcome of many individual competitive interactions can only be understood in terms of macroscopic properties. In essence, the results here echoe the mean field theories that investigate the dynamics of average ecological properties instead of the microscopic components (e.g., McKane et al. 2000). In a philosophical perspective, community ecology has long struggled with analyzing and inferring local determinants of species coexistence from species co-occurrence patterns, so that it was claimed that no universal laws can be derived in the discipline (Lawton 1999). Using different and complementary methods and perspectives, recent research has also shown that species assembly parameter values cannot be unambiguously inferred from species co-occurrences only, even in simple designs where an equilibrium can be reached (Poggiato et al. 2021). Although the roles of high-order competitive interactions and intransivity can lead to species coexistence, the simple view of a single loop of competitive interactions is easily challenged when further interactions and complexity is added (Gallien et al. 2024). But should we put so much emphasis on inferring individual interaction coefficients? In a quest to understand the emerging properties of elementary processes, ecological theory could go forward with a more macroscopic analysis and understanding of species coexistence in many communities. The authors referred several times to an interesting paper from Schaffer (1981), entitled « Ecological abstraction: the consequences of reduced dimensionality in ecological models ». It proposes that estimating individual species competition coefficients is not possible, but that competition can be assessed at the coarser level of organisation, i.e., between ecological guilds. This idea implies that the dimensionality of the competition equations should be greatly reduced to become tractable in practice. Taking together this claim with the results of the present Barbier et al. (2024) paper, it becomes clearer that the nature of competitive interactions can be addressed through « abstracted » quantities, as those of guilds or the moments of the individual competition coefficients (here the average and the standard deviation). Therefore the scope of Barbier et al. (2024) framework goes beyond statistical issues in parameter inference, but question the way we must think and represent the numerous competitive interactions in a simplified and robust way. References Barbier, Matthieu, Jean-François Arnoldi, Guy Bunin, et Michel Loreau. 2018. « Generic assembly patterns in complex ecological communities ». Proceedings of the National Academy of Sciences 115 (9): 2156‑61. https://doi.org/10.1073/pnas.1710352115 | Getting More by Asking for Less: Linking Species Interactions to Species Co-Distributions in Metacommunities | Matthieu Barbier, Guy Bunin, Mathew A. Leibold | <p>AbstractOne of the more difficult challenges in community ecology is inferring species interactions on the basis of patterns in the spatial distribution of organisms. At its core, the problem is that distributional patterns reflect the ‘realize... |  | Biogeography, Community ecology, Competition, Spatial ecology, Metacommunities & Metapopulations, Species distributions, Statistical ecology, Theoretical ecology | François Munoz | 2023-10-21 14:14:16 | View | |
25 Oct 2021

The taxonomic and functional biogeographies of phytoplankton and zooplankton communities across boreal lakesNicolas F St-Gelais, Richard J Vogt, Paul A del Giorgio, Beatrix E Beisner https://doi.org/10.1101/373332The difficult interpretation of species co-distributionRecommended by Dominique Gravel based on reviews by Anthony Maire and Emilie MackeEcology is the study of the distribution of organisms in space and time and their interactions. As such, there is a tradition of studies relating abiotic environmental conditions to species distribution, while another one is concerned by the effects of consumers on the abundance of their resources. Interestingly, joining the dots appears more difficult than it would suggest: eluding the effect of species interactions on distribution remains one of the greatest challenges to elucidate nowadays (Kissling et al. 2012). Theory suggests that yes, species interactions such as predation and competition should influence range limits (Godsoe et al. 2017), but the common intuition among many biogeographers remains that over large areas such as regions and continents, environmental drivers like temperature and precipitation overwhelm their local effects. Answering this question is of primary importance in the context where species are moving around with climate warming. Inconsistencies in food web structure may arise with asynchronized movements of consumers and their resources, leading to a major disruption in regulation and potentially ecosystem functioning. Solving this problem, however, remains very challenging because we have to rely on observational data since experiments are hard to perform at the biogeographical scale. The study of St-Gelais is an interesting step forward to solve this problem. Their main objective was to assess the strength of the association between phytoplankton and zooplankton communities at a large spatial scale, looking at the spatial covariation of both taxonomic and functional composition. To do so, they undertook a massive survey of more than 100 lakes across three regions of the boreal region of Québec. Species and functional composition were recorded, along with a set of abiotic variables. Classic community ecology at this point. The difficulty they faced was to disentangle the multiple causal relationships involved in the distribution of both trophic levels. Teasing apart bottom-up and top-down forces driving the assembly of plankton communities using observational data is not an easy task. On the one hand, both trophic levels could respond to variations in temperature, nutrient availability and dissolved organic carbon. The interpretation is fairly straightforward if the two levels respond to different factors, but the situation is much more complicated when they do respond similarly. There are potentially three possible underlying scenarios. First, the phyto and zooplankton communities may share the same environmental requirements, thereby generating a joint distribution over gradients such as temperature and nutrient availability. Second, the abiotic environment could drive the distribution of the phytoplankton community, which would then propagate up and influence the distribution of the zooplankton community. Alternatively, the abiotic environment could constrain the distribution of the zooplankton, which could then affect the one of phytoplankton. In addition to all of these factors, St-Gelais et al also consider that dispersal may limit the distribution, well aware of previous studies documenting stronger dispersal limitations for zooplankton communities. Unfortunately, there is not a single statistical approach that could be taken from the shelf and used to elucidate drivers of co-distribution. Joint species distribution was once envisioned as a major step forward in this direction (Warton et al. 2015), but there are several limits preventing the direct interpretation that co-occurrence is linked to interactions (Blanchet et al. 2020). Rather, St-Gelais used a variety of multivariate statistics to reveal the structure in their observational data. First, using a Procrustes analysis (a method testing if the spatial variation of one community is correlated to the structure of another community), they found a significant correlation between phytoplankton and zooplankton communities, indicating a taxonomic coupling between the groups. Interestingly, this observation was maintained for functional composition only when interaction-related traits were considered. At this point, these results strongly suggest that interactions are involved in the correlation, but it's hard to decipher between bottom-up and top-down perspectives. A complementary analysis performed with a constrained ordination, per trophic level, provided complementary pieces of information. First observation was that only functional variation was found to be related to the different environmental variables, not taxonomic variation. Despite that trophic levels responded to water quality variables, spatial autocorrelation was more important for zooplankton communities and the two layers appear to respond to different variables. It is impossible with those results to formulate a strong conclusion about whether grazing influence the co-distribution of phytoplankton and zooplankton communities. That's the mere nature of observational data. While there is a strong spatial association between them, there are also diverging responses to the different environmental variables considered. But the contrast between taxonomic and functional composition is nonetheless informative and it seems that beyond the idiosyncrasies of species composition, trait distribution may be more informative and general. Perhaps the most original contribution of this study is the hierarchical approach to analyze the data, combined with the simultaneous analysis of taxonomic and functional distributions. Having access to a vast catalog of multivariate statistical techniques, a careful selection of analyses helps revealing key features in the data, rejecting some hypotheses and accepting others. Hopefully, we will see more and more of such multi-trophic approaches to distribution because it is now clear that the factors driving distribution are much more complicated than anticipated in more traditional analyses of community data. Biodiversity is more than a species list, it is also all of the interactions between them, influencing their distribution and abundance (Jordano 2016). References Blanchet FG, Cazelles K, Gravel D (2020) Co-occurrence is not evidence of ecological interactions. Ecology Letters, 23, 1050–1063. https://doi.org/10.1111/ele.13525 Godsoe W, Jankowski J, Holt RD, Gravel D (2017) Integrating Biogeography with Contemporary Niche Theory. Trends in Ecology & Evolution, 32, 488–499. https://doi.org/10.1016/j.tree.2017.03.008 Jordano P (2016) Chasing Ecological Interactions. PLOS Biology, 14, e1002559. https://doi.org/10.1371/journal.pbio.1002559 Kissling WD, Dormann CF, Groeneveld J, Hickler T, Kühn I, McInerny GJ, Montoya JM, Römermann C, Schiffers K, Schurr FM, Singer A, Svenning J-C, Zimmermann NE, O’Hara RB (2012) Towards novel approaches to modelling biotic interactions in multispecies assemblages at large spatial extents. Journal of Biogeography, 39, 2163–2178. https://doi.org/10.1111/j.1365-2699.2011.02663.x St-Gelais NF, Vogt RJ, Giorgio PA del, Beisner BE (2021) The taxonomic and functional biogeographies of phytoplankton and zooplankton communities across boreal lakes. bioRxiv, 373332, ver. 4 peer-reviewed and recommended by Peer community in Ecology. https://doi.org/10.1101/373332 Warton DI, Blanchet FG, O’Hara RB, Ovaskainen O, Taskinen S, Walker SC, Hui FKC (2015) So Many Variables: Joint Modeling in Community Ecology. Trends in Ecology & Evolution, 30, 766–779. https://doi.org/10.1016/j.tree.2015.09.007 Wisz MS, Pottier J, Kissling WD, Pellissier L, Lenoir J, Damgaard CF, Dormann CF, Forchhammer MC, Grytnes J-A, Guisan A, Heikkinen RK, Høye TT, Kühn I, Luoto M, Maiorano L, Nilsson M-C, Normand S, Öckinger E, Schmidt NM, Termansen M, Timmermann A, Wardle DA, Aastrup P, Svenning J-C (2013) The role of biotic interactions in shaping distributions and realised assemblages of species: implications for species distribution modelling. Biological Reviews, 88, 15–30. https://doi.org/10.1111/j.1469-185X.2012.00235.x | The taxonomic and functional biogeographies of phytoplankton and zooplankton communities across boreal lakes | Nicolas F St-Gelais, Richard J Vogt, Paul A del Giorgio, Beatrix E Beisner | <p>Strong trophic interactions link primary producers (phytoplankton) and consumers (zooplankton) in lakes. However, the influence of such interactions on the biogeographical distribution of the taxa and functional traits of planktonic organ... |  | Biogeography, Community ecology, Species distributions | Dominique Gravel | 2018-07-24 15:01:51 | View | |
04 Apr 2023
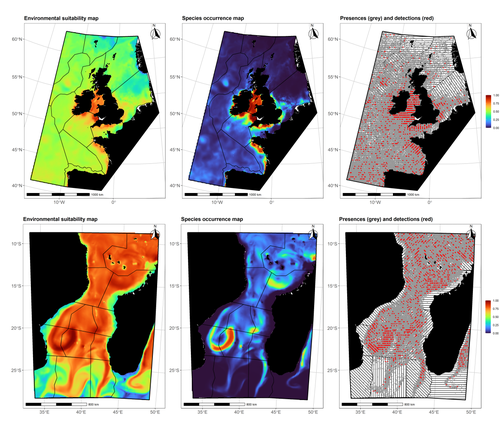
Data stochasticity and model parametrisation impact the performance of species distribution models: insights from a simulation studyCharlotte Lambert, Auriane Virgili https://doi.org/10.1101/2023.01.17.524386Species Distribution Models: the delicate balance between signal and noiseRecommended by Timothée Poisot based on reviews by Alejandra Zarzo Arias and 1 anonymous reviewer based on reviews by Alejandra Zarzo Arias and 1 anonymous reviewer
Species Distribution Models (SDMs) are one of the most commonly used tools to predict where species are, where they may be in the future, and, at times, what are the variables driving this prediction. As such, applying an SDM to a dataset is akin to making a bet: that the known occurrence data are informative, that the resolution of predictors is adequate vis-à-vis the scale at which their impact is expressed, and that the model will adequately capture the shape of the relationships between predictors and predicted occurrence. In this contribution, Lambert & Virgili (2023) perform a comprehensive assessment of different sources of complications to this process, using replicated simulations of two synthetic species. Their experimental process is interesting, in that both the data generation and the data analysis stick very close to what would happen in "real life". The use of synthetic species is particularly relevant to the assessment of SDM robustness, as they enable the design of species for which the shape of the relationship is given: in short, we know what the model should capture, and can evaluate the model performance against a ground truth that lacks uncertainty. Any simulation study is limited by the assumptions established by the investigators; when it comes to spatial data, the "shape" of the landscape, both in terms of auto-correlation and in where the predictors are available. Lambert & Virgili (2023) nicely circumvent these issues by simulating synthetic species against the empirical distribution of predictors; in other words, the species are synthetic, but the environment for which the prediction is made is real. This is an important step forward when compared to the use of e.g. neutral landscapes (With 1997), which can have statistical properties that are not representative of natural landscapes (see e.g. Halley et al., 2004). A striking point in the study by Lambert & Virgili (2023) is that they reveal a deep, indeed deeper than expected, stochasticity in SDMs; whether this is true in all models remains an open question, but does not invalidate their recommendation to the community: the interpretation of outcomes is a delicate exercise, especially because measures that inform on the goodness of the model fit do not capture the predictive quality of the model outputs. This preprint is both a call to more caution, and a call to more curiosity about the complex behavior of SDMs, while also providing a sensible template to perform future analyses of the potential issues with predictive models.
Halley, J. M., et al. (2004) “Uses and Abuses of Fractal Methodology in Ecology: Fractal Methodology in Ecology.” Ecology Letters, vol. 7, no. 3, pp. 254–71. https://doi.org/10.1111/j.1461-0248.2004.00568.x. Lambert, Charlotte, and Auriane Virgili (2023). Data Stochasticity and Model Parametrisation Impact the Performance of Species Distribution Models: Insights from a Simulation Study. bioRxiv, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.01.17.524386 With, Kimberly A. (1997) “The Application of Neutral Landscape Models in Conservation Biology. Aplicacion de Modelos de Paisaje Neutros En La Biologia de La Conservacion.” Conservation Biology, vol. 11, no. 5, pp. 1069–80. https://doi.org/10.1046/j.1523-1739.1997.96210.x. | Data stochasticity and model parametrisation impact the performance of species distribution models: insights from a simulation study | Charlotte Lambert, Auriane Virgili | <p>Species distribution models (SDM) are widely used to describe and explain how species relate to their environment, and predict their spatial distributions. As such, they are the cornerstone of most of spatial planning efforts worldwide. SDM can... |  | Biogeography, Habitat selection, Macroecology, Marine ecology, Spatial ecology, Metacommunities & Metapopulations, Species distributions, Statistical ecology | Timothée Poisot | 2023-01-20 09:43:51 | View | |
31 Oct 2022

Ten simple rules for working with high resolution remote sensing dataAdam L. Mahood, Maxwell Benjamin Joseph, Anna Spiers, Michael J. Koontz, Nayani Ilangakoon, Kylen Solvik, Nathan Quarderer, Joe McGlinchy, Victoria Scholl, Lise St. Denis, Chelsea Nagy, Anna Braswell, Matthew W. Rossi, Lauren Herwehe, Leah wasser, Megan Elizabeth Cattau, Virginia Iglesias, Fangfang Yao, Stefan Leyk, Jennifer Balch https://doi.org/10.31219/osf.io/kehqzPreventing misuse of high-resolution remote sensing dataRecommended by Eric Goberville based on reviews by Jane Wyngaard and 1 anonymous reviewer based on reviews by Jane Wyngaard and 1 anonymous reviewer
To observe, characterise, identify, understand, predict... This is the approach that researchers follow every day. This sequence is tirelessly repeated as the biological model, the targeted ecosystem and/or the experimental, environmental or modelling conditions change. This way of proceeding is essential in a world of rapid change in response to the frenetic pace of intensifying pressures and forcings that impact ecosystems. To better understand our Earth and the dynamics of its components, to map ecosystems and diversity patterns, and to identify changes, humanity had to demonstrate inventiveness and defy gravity. Gustave Hermite and Georges Besançon were the first to launch aloft balloons equipped with radio transmitters, making possible the transmission of meteorological data to observers in real time [1]. The development of aviation in the middle of the 20th century constituted a real leap forward for the frequent acquisition of aerial observations, leading to a significant improvement in weather forecasting models. The need for systematic collection of data as holistic as possible – an essential component for the observation of complex biological systems - has resulted in pushing the limits of technological prowess. The conquest of space and the concurrent development of satellite observations has largely contributed to the collection of a considerable mass of data, placing our Earth under the "macroscope" - a concept introduced to ecology in the early 1970s by Howard T. Odum (see [2]), and therefore allowing researchers to move towards a better understanding of ecological systems, deterministic and stochastic patterns … with the ultimate goal of improving management actions [2,3]. Satellite observations have been carried out for nearly five decades now [3] and have greatly contributed to a better qualitative and quantitative understanding of the functioning of our planet, its diversity, its climate... and to a better anticipation of possible future changes (e.g., [4-7]). This access to rich and complex sources of information, for which both spatial and temporal resolutions are increasingly fine, results in the implementation of increasingly complex computation-based analyses, in order to meet the need for a better understanding of ecological mechanisms and processes, and their possible changes. Steven Levitt stated that "Data is one of the most powerful mechanisms for telling stories". This is so true … Data should not be used as a guide to thinking and a critical judgment at each stage of the data exploitation process should not be neglected. This is what Mahood et al. [8] rightly remind us in their article "Ten simple rules for working with high-resolution remote sensing data" in which they provide the fundamentals to consider when working with data of this nature, a still underutilized resource in several topics, such as conservation biology [3]. In this unconventional article, presented in a pedagogical way, the authors remind different generations of readers how satellite data should be handled and processed. The authors aim to make the readers aware of the most frequent pitfalls encouraging them to use data adapted to their original question, the most suitable tools/methods/procedures, to avoid methodological overkill, and to ensure both ethical use of data and transparency in the research process. While access to high-resolution data is increasingly easy thanks to the implementation of dedicated platforms [4], and because of the development of easy-to-use processing software and pipelines, it is important to take the time to recall some of the essential rules and guidelines for managing them, from new users with little or no experience who will find in this article the recommendations, resources and advice necessary to start exploiting remote sensing data, to more experienced researchers. References [1] Jeannet P, Philipona R, and Richner H (2016). 8 Swiss upper-air balloon soundings since 1902. In: Willemse S, Furger M (2016) From weather observations to atmospheric and climate sciences in Switzerland: Celebrating 100 years of the Swiss Society for Meteorology. vdf Hochschulverlag AG. [2] Odum HT (2007) Environment, Power, and Society for the Twenty-First Century: The Hierarchy of Energy. Columbia University Press. [3] Boyle SA, Kennedy CM, Torres J, Colman K, Pérez-Estigarribia PE, Sancha NU de la (2014) High-Resolution Satellite Imagery Is an Important yet Underutilized Resource in Conservation Biology. PLOS ONE, 9, e86908. https://doi.org/10.1371/journal.pone.0086908 [4] Le Traon P-Y, Antoine D, Bentamy A, Bonekamp H, Breivik LA, Chapron B, Corlett G, Dibarboure G, DiGiacomo P, Donlon C, Faugère Y, Font J, Girard-Ardhuin F, Gohin F, Johannessen JA, Kamachi M, Lagerloef G, Lambin J, Larnicol G, Le Borgne P, Leuliette E, Lindstrom E, Martin MJ, Maturi E, Miller L, Mingsen L, Morrow R, Reul N, Rio MH, Roquet H, Santoleri R, Wilkin J (2015) Use of satellite observations for operational oceanography: recent achievements and future prospects. Journal of Operational Oceanography, 8, s12–s27. https://doi.org/10.1080/1755876X.2015.1022050 [5] Turner W, Rondinini C, Pettorelli N, Mora B, Leidner AK, Szantoi Z, Buchanan G, Dech S, Dwyer J, Herold M, Koh LP, Leimgruber P, Taubenboeck H, Wegmann M, Wikelski M, Woodcock C (2015) Free and open-access satellite data are key to biodiversity conservation. Biological Conservation, 182, 173–176. https://doi.org/10.1016/j.biocon.2014.11.048 [6] Melet A, Teatini P, Le Cozannet G, Jamet C, Conversi A, Benveniste J, Almar R (2020) Earth Observations for Monitoring Marine Coastal Hazards and Their Drivers. Surveys in Geophysics, 41, 1489–1534. https://doi.org/10.1007/s10712-020-09594-5 [7] Zhao Q, Yu L, Du Z, Peng D, Hao P, Zhang Y, Gong P (2022) An Overview of the Applications of Earth Observation Satellite Data: Impacts and Future Trends. Remote Sensing, 14, 1863. https://doi.org/10.3390/rs14081863 [8] Mahood AL, Joseph MB, Spiers A, Koontz MJ, Ilangakoon N, Solvik K, Quarderer N, McGlinchy J, Scholl V, Denis LS, Nagy C, Braswell A, Rossi MW, Herwehe L, Wasser L, Cattau ME, Iglesias V, Yao F, Leyk S, Balch J (2021) Ten simple rules for working with high resolution remote sensing data. OSFpreprints, ver. 6 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.31219/osf.io/kehqz | Ten simple rules for working with high resolution remote sensing data | Adam L. Mahood, Maxwell Benjamin Joseph, Anna Spiers, Michael J. Koontz, Nayani Ilangakoon, Kylen Solvik, Nathan Quarderer, Joe McGlinchy, Victoria Scholl, Lise St. Denis, Chelsea Nagy, Anna Braswell, Matthew W. Rossi, Lauren Herwehe, Leah wasser,... | <p>Researchers in Earth and environmental science can extract incredible value from high-resolution (sub-meter, sub-hourly or hyper-spectral) remote sensing data, but these data can be difficult to use. Correct, appropriate and competent use of su... |  | Biogeography, Landscape ecology, Macroecology, Spatial ecology, Metacommunities & Metapopulations, Terrestrial ecology | Eric Goberville | 2021-10-19 21:41:22 | View | |
30 Mar 2020
Environmental variables determining the distribution of an avian parasite: the case of the Philornis torquans complex (Diptera: Muscidae) in South AmericaPablo F. Cuervo, Alejandro Percara, Lucas Monje, Pablo M. Beldomenico, Martín A. Quiroga https://doi.org/10.1101/839589Catching the fly in dystopian timesRecommended by Rodrigo Medel based on reviews by 4 anonymous reviewersHost-parasite interactions are ubiquitous on Earth. They are present in almost every conceivable ecosystem and often result from a long history of antagonist coevolution [1,2]. Recent studies on climate change have revealed, however, that modification of abiotic variables are often accompanied by shifts in the distributional range of parasites to habitats far beyond their original geographical distribution, creating new interactions in novel habitats with unpredictable consequences for host community structure and organization [3,4]. This situation may be especially critical for endangered host species having small population abundance and restricted distribution range. The infestation of bird species with larvae of the muscid fly genus Philornis is a case in point. At least 250 bird species inhabiting mostly Central and South America are infected by Philornis flies [5,6]. Fly larval development occurs in bird faeces, nesting material, or inside nestlings, affecting the development and nestling survival. References [1] Thompson JN (1994) The Coevolutionary Process. University of Chicago Press. | Environmental variables determining the distribution of an avian parasite: the case of the Philornis torquans complex (Diptera: Muscidae) in South America | Pablo F. Cuervo, Alejandro Percara, Lucas Monje, Pablo M. Beldomenico, Martín A. Quiroga | <p>*Philornis* flies are the major cause of myasis in altricial nestlings of neotropical birds. Its impact ranges from subtle to lethal, being of major concern in endangered bird species with geographically-restricted, fragmented and small-sized p... |  | Biogeography, Macroecology, Parasitology, Species distributions | Rodrigo Medel | 2019-11-26 21:31:33 | View | |
29 Jun 2024
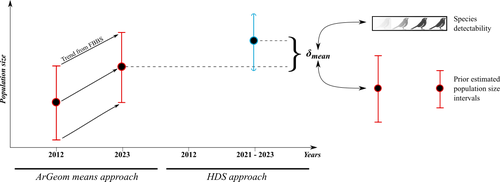
Reassessment of French breeding bird population sizes using citizen science and accounting for species detectabilityJean Nabias, Luc Barbaro, Benoit Fontaine, Jérémy Dupuy, Laurent Couzi, Clément Vallé, Romain Lorrillière https://hal.science/hal-04478371Reassessment of French breeding bird population sizes: from citizen science observations to nationwide estimatesRecommended by Nigel Yoccoz based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Estimating populations size of widespread, common species in a relatively large and heterogeneous country like France is difficult for several reasons, from having a sample covering well the diverse ecological gradients to accounting for detectability, the fact that absence of a species may represent a false negative, the species being present but not detected. Bird communities have been the focus of a very large number of studies, with some countries like the UK having long traditions of monitoring both common and rare species. Nabias et al. use a large, structured citizen science project to provide new estimates of common bird species, accounting for detectability and using different habitat and climate covariates to extrapolate abundance to non-sampled areas. About 2/3 of the species had estimates higher than what would have been expected using a previous attempt at estimating population size based in part on expert knowledge and projected using estimates of trends to the period covered by the citizen science sampling. Some species showed large differences between the two estimates, which could be in part explained by accounting for detectability. This paper uses what is called model-based inference (as opposed to design-based inference, that uses the design to make inferences about the whole population; Buckland et al. 2000), both in terms of detectability and habitat suitability. The estimates obtained depend on how well the model components approximate the underlying processes, which in a complex dataset like this one is not easy to assess. But it clearly shows that detectability may have substantial implications for the population size estimates. This is of course not new but has rarely been done at this scale and using a large sample obtained on many species. Interesting further work could focus on testing the robustness of the model-based approach by for example sampling new plots and compare the expected values to the observed values. Such a sampling could be stratified to maximize the discrimination between expected low and high abundances, at least for species where the estimates might be considered as uncertain, or for which estimating population sizes is deemed important. References Buckland, S. T., Goudie, I. B. J., & Borchers, D. L. (2000). Wildlife Population Assessment: Past Developments and Future Directions. Biometrics, 56(1), 1-12. https://doi.org/10.1111/j.0006-341X.2000.00001.x Nabias, J., Barbaro, L., Fontaine, B., Dupuy, J., Couzi, L., et al. (2024) Reassessment of French breeding bird population sizes using citizen science and accounting for species detectability. HAL, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://hal.science/hal-04478371 | Reassessment of French breeding bird population sizes using citizen science and accounting for species detectability | Jean Nabias, Luc Barbaro, Benoit Fontaine, Jérémy Dupuy, Laurent Couzi, Clément Vallé, Romain Lorrillière | <p style="text-align: justify;">Higher efficiency in large-scale and long-term biodiversity monitoring can be obtained through the use of Essential Biodiversity Variables, among which species population sizes provide key data for conservation prog... |  | Biogeography, Macroecology, Spatial ecology, Metacommunities & Metapopulations, Species distributions, Statistical ecology | Nigel Yoccoz | 2024-02-26 18:10:27 | View | |
12 Jun 2019
Environmental heterogeneity drives tsetse fly population dynamics and controlCecilia H, Arnoux S, Picault S, Dicko A, Seck MT, Sall B, Bassene M, Vreysen M, Pagabeleguem S, Bance A, Bouyer J, Ezanno P https://doi.org/10.1101/493650Modeling jointly landscape complexity and environmental heterogeneity to envision new strategies for tsetse flies controlRecommended by Benjamin Roche based on reviews by Timothée Vergne and 1 anonymous reviewerToday, understanding spatio-temporal dynamics of pathogens is pivotal to understand their transmission and controlling them. First, understanding this dynamics can reveal the ecology of their transmission [1]. Indeed, such knowledge, based on data that are quite easy to access, can shed light on transmission modes, which could rely on different animal species that can be spatially distributed in a non-uniform way [2]. This is especially true for pathogens with complex life-cycles, despite that investigating such dynamics is very challenging and rely mostly on mathematical models. References [1] Grenfell, B. T., Bjørnstad, O. N., & Kappey, J. (2001). Travelling waves and spatial hierarchies in measles epidemics. Nature, 414(6865), 716-723. doi: 10.1038/414716a | Environmental heterogeneity drives tsetse fly population dynamics and control | Cecilia H, Arnoux S, Picault S, Dicko A, Seck MT, Sall B, Bassene M, Vreysen M, Pagabeleguem S, Bance A, Bouyer J, Ezanno P | <p>A spatially and temporally heterogeneous environment may lead to unexpected population dynamics. Knowledge still is needed on which of the local environment properties favour population maintenance at larger scale. For pathogen vectors, such as... |  | Biological control, Population ecology, Spatial ecology, Metacommunities & Metapopulations | Benjamin Roche | 2018-12-14 12:13:39 | View | |
05 Apr 2022
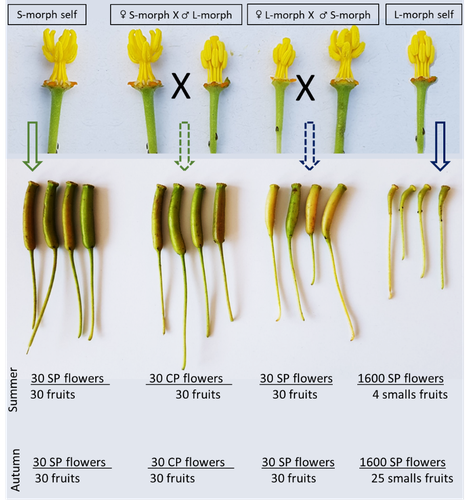
Late-acting self-incompatible system, preferential allogamy and delayed selfing in the heterostylous invasive populations of Ludwigia grandiflora subsp. hexapetalaLuis O. Portillo Lemus, Maryline Harang, Michel Bozec, Jacques Haury, Solenn Stoeckel, Dominique Barloy https://doi.org/10.1101/2021.07.15.452457Water primerose (Ludwigia grandiflora subsp. hexapetala) auto- and allogamy: an ecological perspectiveRecommended by Antoine Vernay based on reviews by Juan Arroyo, Emiliano Mora-Carrera and 1 anonymous reviewer based on reviews by Juan Arroyo, Emiliano Mora-Carrera and 1 anonymous reviewer
Invasive plant species are widely studied by the ecologist community, especially in wetlands. Indeed, alien plants are considered one of the major threats to wetland biodiversity (Reid et al., 2019). Ludwigia grandiflora subsp. hexapetala (Hook. & Arn.) G.L.Nesom & Kartesz, 2000 (Lgh) is one of them and has received particular attention for a long time (Hieda et al., 2020; Thouvenot, Haury, & Thiebaut, 2013). The ecology of this invasive species and its effect on its biotic and abiotic environment has been studied in previous works. Different processes were demonstrated to explain their invasibility such as allelopathic interference (Dandelot et al., 2008), resource competition (Gérard et al., 2014), and high phenotypic plasticity (Thouvenot, Haury, & Thiébaut, 2013), to cite a few of them. However, although vegetative reproduction is a well-known invasive process for alien plants like Lgh (Glover et al., 2015), the sexual reproduction of this species is still unclear and may help to understand the Lgh population dynamics. Portillo Lemus et al. (2021) showed that two floral morphs of Lgh co-exist in natura, involving self-compatibility for short-styled phenotype and self-incompatibility for long-styled phenotype processes. This new article (Portillo Lemus et al., 2022) goes further and details the underlying mechanisms of the sexual reproduction of the two floral morphs. Complementing their previous study, the authors have described a late self-incompatible process associated with the long-styled morph, which authorized a small proportion of autogamy. Although this represents a small fraction of the L-morph reproduction, it may have a considerable impact on the L-morph population dynamics. Indeed, authors report that “floral morphs are mostly found in allopatric monomorphic populations (i.e., exclusively S-morph or exclusively L-morph populations)” with a large proportion of L-morph populations compared to S-morph populations in the field. It may seem counterintuitive as L-morph mainly relies on cross-fecundation. Results show that L-morph autogamy mainly occurs in the fall, late in the reproduction season. Therefore, the reproduction may be ensured if no exogenous pollen reaches the stigma of L-morph individuals. It partly explains the large proportion of L-morph populations in the field. Beyond the description of late-acting self-incompatibility, which makes the Onagraceae a third family of Myrtales with this reproductive adaptation, the study raises several ecological questions linked to the results presented in the article. First, it seems that even if autogamy is possible, Lgh would favour allogamy, even in S-morph, through the faster development of pollen tubes from other individuals. This may confer an adaptative and evolutive advantage for the Lgh, increasing its invasive potential. The article shows this faster pollen tube development in S-morph but does not test the evolutive consequences. It is an interesting perspective for future research. It would also be interesting to describe cellular processes which recognize and then influence the speed of the pollen tube. Second, the importance of sexual reproduction vs vegetative reproduction would also provide information on the benefits of sexual dimorphism within populations. For instance, how fruit production increases the dispersal potential of Lgh would help to understand Lgh population dynamics and to propose adapted management practices (Delbart et al., 2013; Meisler, 2009). To conclude, the study proposes a morphological, reproductive and physiological description of the Lgh sexual reproduction process. However, underlying ecological questions are well included in the article and the ecophysiological results enlighten some questions about the role of sexual reproduction in the invasiveness of Lgh. I advise the reader to pay attention to the reviewers’ comments; the debates were very constructive and, thanks to the great collaboration with the authorship, lead to an interesting paper about Lgh reproduction and with promising perspectives in ecology and invasion ecology. References Dandelot S, Robles C, Pech N, Cazaubon A, Verlaque R (2008) Allelopathic potential of two invasive alien Ludwigia spp. Aquatic Botany, 88, 311–316. https://doi.org/10.1016/j.aquabot.2007.12.004 Delbart E, Mahy G, Monty A (2013) Efficacité des méthodes de lutte contre le développement de cinq espèces de plantes invasives amphibies : Crassula helmsii, Hydrocotyle ranunculoides, Ludwigia grandiflora, Ludwigia peploides et Myriophyllum aquaticum (synthèse bibliographique). BASE, 17, 87–102. https://popups.uliege.be/1780-4507/index.php?id=9586 Gérard J, Brion N, Triest L (2014) Effect of water column phosphorus reduction on competitive outcome and traits of Ludwigia grandiflora and L. peploides, invasive species in Europe. Aquatic Invasions, 9, 157–166. https://doi.org/10.3391/ai.2014.9.2.04 Glover R, Drenovsky RE, Futrell CJ, Grewell BJ (2015) Clonal integration in Ludwigia hexapetala under different light regimes. Aquatic Botany, 122, 40–46. https://doi.org/10.1016/j.aquabot.2015.01.004 Hieda S, Kaneko Y, Nakagawa M, Noma N (2020) Ludwigia grandiflora (Michx.) Greuter & Burdet subsp. hexapetala (Hook. & Arn.) G. L. Nesom & Kartesz, an Invasive Aquatic Plant in Lake Biwa, the Largest Lake in Japan. Acta Phytotaxonomica et Geobotanica, 71, 65–71. https://doi.org/10.18942/apg.201911 Meisler J (2009) Controlling Ludwigia hexaplata in Northern California. Wetland Science and Practice, 26, 15–19. https://doi.org/10.1672/055.026.0404 Portillo Lemus LO, Harang M, Bozec M, Haury J, Stoeckel S, Barloy D (2022) Late-acting self-incompatible system, preferential allogamy and delayed selfing in the heteromorphic invasive populations of Ludwigia grandiflora subsp. hexapetala. bioRxiv, 2021.07.15.452457, ver. 4 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2021.07.15.452457 Portillo Lemus LO, Bozec M, Harang M, Coudreuse J, Haury J, Stoeckel S, Barloy D (2021) Self-incompatibility limits sexual reproduction rather than environmental conditions in an invasive water primrose. Plant-Environment Interactions, 2, 74–86. https://doi.org/10.1002/pei3.10042 Reid AJ, Carlson AK, Creed IF, Eliason EJ, Gell PA, Johnson PTJ, Kidd KA, MacCormack TJ, Olden JD, Ormerod SJ, Smol JP, Taylor WW, Tockner K, Vermaire JC, Dudgeon D, Cooke SJ (2019) Emerging threats and persistent conservation challenges for freshwater biodiversity. Biological Reviews, 94, 849–873. https://doi.org/10.1111/brv.12480 Thouvenot L, Haury J, Thiebaut G (2013) A success story: water primroses, aquatic plant pests. Aquatic Conservation: Marine and Freshwater Ecosystems, 23, 790–803. https://doi.org/10.1002/aqc.2387 Thouvenot L, Haury J, Thiébaut G (2013) Seasonal plasticity of Ludwigia grandiflora under light and water depth gradients: An outdoor mesocosm experiment. Flora - Morphology, Distribution, Functional Ecology of Plants, 208, 430–437. https://doi.org/10.1016/j.flora.2013.07.004 | Late-acting self-incompatible system, preferential allogamy and delayed selfing in the heterostylous invasive populations of Ludwigia grandiflora subsp. hexapetala | Luis O. Portillo Lemus, Maryline Harang, Michel Bozec, Jacques Haury, Solenn Stoeckel, Dominique Barloy | <p style="text-align: justify;">Breeding system influences local population genetic structure, effective size, offspring fitness and functional variation. Determining the respective importance of self- and cross-fertilization in hermaphroditic flo... |  | Biological invasions, Botany, Freshwater ecology, Pollination | Antoine Vernay | 2021-07-16 09:53:50 | View | |
16 Jun 2023
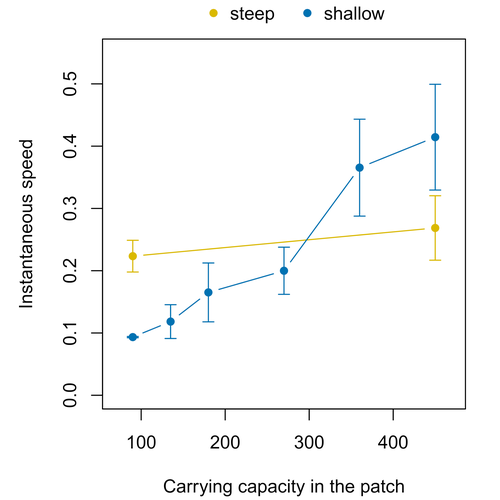
Colonisation debt: when invasion history impacts current range expansionThibaut Morel-Journel, Marjorie Haond, Lana Duan, Ludovic Mailleret, Elodie Vercken https://doi.org/10.1101/2022.11.13.516255Combining stochastic models and experiments to understand dispersal in heterogeneous environmentsRecommended by Joaquín Hortal based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Dispersal is a key element of the natural dynamics of meta-communities, and plays a central role in the success of populations colonizing new landscapes. Understanding how demographic processes may affect the speed at which alien species spread through environmentally-heterogeneous habitat fragments is therefore of key importance to manage biological invasions. This requires studying together the complex interplay of dispersal and population processes, two inextricably related phenomena that can produce many possible outcomes. Stochastic models offer an opportunity to describe this kind of process in a meaningful way, but to ensure that they are realistic (sensu Levins 1966) it is also necessary to combine model simulations with empirical data (Snäll et al. 2007). Morel-Journel et al. (2023) put together stochastic models and experimental data to study how population density may affect the speed at which alien species spread through a heterogeneous landscape. They do it by focusing on what they call ‘colonisation debt’, which is merely the impact that population density at the invasion front may have on the speed at which the species colonizes patches of different carrying capacities. They investigate this issue through two largely independent approaches. First, a stochastic model of dispersal throughout the patches of a linear, 1-dimensional landscape, which accounts for different degrees of density-dependent growth. And second, a microcosm experiment of a parasitoid wasp colonizing patches with different numbers of host eggs. In both cases, they compare the velocity of colonization of patches with lower or higher carrying capacity than the previous one (i.e. what they call upward or downward gradients). Their results show that density-dependent processes influence the speed at which new fragments are colonized is significantly reduced by positive density dependence. When either population growth or dispersal rate depend on density, colonisation debt limits the speed of invasion, which turns out to be dependent on the strength and direction of the gradient between the conditions of the invasion front, and the newly colonized patches. Although this result may be quite important to understand the meta-population dynamics of dispersing species, it is important to note that in their study the environmental differences between patches do not take into account eventual shifts in the scenopoetic conditions (i.e. the values of the environmental parameters to which species niches’ respond to; Hutchinson 1978, see also Soberón 2007). Rather, differences arise from variations in the carrying capacity of the patches that are consecutively invaded, both in the in silico and microcosm experiments. That is, they account for potential differences in the size or quality of the invaded fragments, but not on the costs of colonizing fragments with different environmental conditions, which may also determine invasion speed through niche-driven processes. This aspect can be of particular importance in biological invasions or under climate change-driven range shifts, when adaptation to new environments is often required (Sakai et al. 2001; Whitney & Gabler 2008; Hill et al. 2011). The expansion of geographical distribution ranges is the result of complex eco-evolutionary processes where meta-community dynamics and niche shifts interact in a novel physical space and/or environment (see, e.g., Mestre et al. 2020). Here, the invasibility of native communities is determined by niche variations and how similar are the traits of alien and native species (Hui et al. 2023). Within this context, density-dependent processes will build upon and heterogeneous matrix of native communities and environments (Tischendorf et al. 2005), to eventually determine invasion success. What the results of Morel-Journel et al. (2023) show is that, when the invader shows density dependence, the invasion process can be slowed down by variations in the carrying capacity of patches along the dispersal front. This can be particularly useful to manage biological invasions; ongoing invasions can be at least partially controlled by manipulating the size or quality of the patches that are most adequate to the invader, controlling host populations to reduce carrying capacity. But further, landscape manipulation of such kind could be used in a preventive way, to account in advance for the effects of the introduction of alien species for agricultural exploitation or biological control, thereby providing an additional safeguard to practices such as the introduction of parasitoids to control plagues. These practical aspects are certainly worth exploring further, together with a more explicit account of the influence of the abiotic conditions and the characteristics of the invaded communities on the success and speed of biological invasions. REFERENCES Hill, J.K., Griffiths, H.M. & Thomas, C.D. (2011) Climate change and evolutionary adaptations at species' range margins. Annual Review of Entomology, 56, 143-159. https://doi.org/10.1146/annurev-ento-120709-144746 Hui, C., Pyšek, P. & Richardson, D.M. (2023) Disentangling the relationships among abundance, invasiveness and invasibility in trait space. npj Biodiversity, 2, 13. https://doi.org/10.1038/s44185-023-00019-1 Hutchinson, G.E. (1978) An introduction to population biology. Yale University Press, New Haven, CT. Levins, R. (1966) The strategy of model building in population biology. American Scientist, 54, 421-431. Mestre, A., Poulin, R. & Hortal, J. (2020) A niche perspective on the range expansion of symbionts. Biological Reviews, 95, 491-516. https://doi.org/10.1111/brv.12574 Morel-Journel, T., Haond, M., Duan, L., Mailleret, L. & Vercken, E. (2023) Colonisation debt: when invasion history impacts current range expansion. bioRxiv, 2022.11.13.516255, ver. 3 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2022.11.13.516255 Snäll, T., B. O'Hara, R. & Arjas, E. (2007) A mathematical and statistical framework for modelling dispersal. Oikos, 116, 1037-1050. https://doi.org/10.1111/j.0030-1299.2007.15604.x Sakai, A.K., Allendorf, F.W., Holt, J.S., Lodge, D.M., Molofsky, J., With, K.A., Baughman, S., Cabin, R.J., Cohen, J.E., Ellstrand, N.C., McCauley, D.E., O'Neil, P., Parker, I.M., Thompson, J.N. & Weller, S.G. (2001) The population biology of invasive species. Annual Review of Ecology and Systematics, 32, 305-332. https://doi.org/10.1146/annurev.ecolsys.32.081501.114037 Soberón, J. (2007) Grinnellian and Eltonian niches and geographic distributions of species. Ecology Letters, 10, 1115-1123. https://doi.org/10.1111/j.1461-0248.2007.01107.x Tischendorf, L., Grez, A., Zaviezo, T. & Fahrig, L. (2005) Mechanisms affecting population density in fragmented habitat. Ecology and Society, 10, 7. https://doi.org/10.5751/ES-01265-100107 Whitney, K.D. & Gabler, C.A. (2008) Rapid evolution in introduced species, 'invasive traits' and recipient communities: challenges for predicting invasive potential. Diversity and Distributions, 14, 569-580. https://doi.org/10.1111/j.1472-4642.2008.00473.x | Colonisation debt: when invasion history impacts current range expansion | Thibaut Morel-Journel, Marjorie Haond, Lana Duan, Ludovic Mailleret, Elodie Vercken | <p>Demographic processes that occur at the local level, such as positive density dependence in growth or dispersal, are known to shape population range expansion, notably by linking carrying capacity to invasion speed. As a result of these process... |  | Biological invasions, Colonization, Dispersal & Migration, Experimental ecology, Landscape ecology, Population ecology, Spatial ecology, Metacommunities & Metapopulations, Theoretical ecology | Joaquín Hortal | Anonymous, Anonymous | 2022-11-16 15:52:08 | View |
MANAGING BOARD
Julia Astegiano
Tim Coulson
Anna Eklof
Dominique Gravel
François Massol
Ben Phillips
Cyrille Violle










