Direct submissions to PCI Ecology from bioRxiv.org are possible using the B2J service
Latest recommendations

| Id | Title * ▲ | Authors * | Abstract * | Picture * | Thematic fields * | Recommender | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
01 Oct 2023
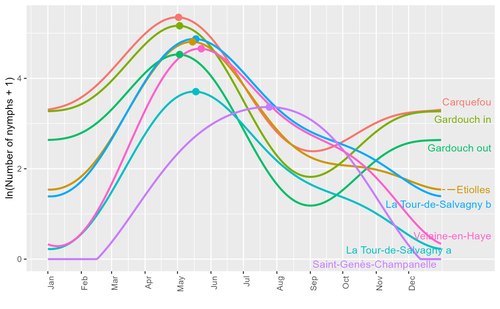
Seasonality of host-seeking Ixodes ricinus nymph abundance in relation to climateThierry Hoch, Aurélien Madouasse, Maude Jacquot, Phrutsamon Wongnak, Fréderic Beugnet, Laure Bournez, Jean-François Cosson, Frédéric Huard, Sara Moutailler, Olivier Plantard, Valérie Poux, Magalie René-Martellet, Muriel Vayssier-Taussat, Hélène Verheyden, Gwenaёl Vourc’h, Karine Chalvet-Monfray, Albert Agoulon https://doi.org/10.1101/2022.07.25.501416Assessing seasonality of tick abundance in different climatic regionsRecommended by Nigel Yoccoz based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Tick-borne pathogens are considered as one of the major threats to public health – Lyme borreliosis being a well-known example of such disease. Global change – from climate change to changes in land use or invasive species – is playing a role in the increased risk associated with these pathogens. An important aspect of our knowledge of ticks and their associated pathogens is seasonality – one component being the phenology of within-year tick occurrences. This is important both in terms of health risk – e.g., when is the risk of encountering ticks high – and ecological understanding, as tick dynamics may depend on the weather as well as different hosts with their own dynamics and habitat use. Hoch et al. (2023) provide a detailed data set on the phenology of one species of tick, Ixodes ricinus, in 6 different locations of France. Whereas relatively cool sites showed a clear peak in spring-summer, warmer sites showed in addition relatively high occurrences in fall-winter, with a minimum in late summer-early fall. Such results add robust data to the existing evidence of the importance of local climatic patterns for explaining tick phenology. Recent analyses have shown that the phenology of Lyme borreliosis has been changing in northern Europe in the last 25 years, with seasonal peaks in cases occurring now 6 weeks earlier (Goren et al. 2023). The study by Hoch et al. (2023) is of too short duration to establish temporal changes in phenology (“only” 8 years, 2014-2021, see also Wongnak et al 2021 for some additional analyses; given the high year-to-year variability in weather, establishing phenological changes often require longer time series), and further work is needed to get better estimates of these changes and relate them to climate, land-use, and host density changes. Moreover, the phenology of ticks may also be related to species-specific tick phenology, and different tick species do not respond to current changes in identical ways (see for example differences between the two Ixodes species in Finland; Uusitalo et al. 2022). An efficient surveillance system requires therefore an adaptive monitoring design with regard to the tick species as well as the evolving causes of changes. References Goren, A., Viljugrein, H., Rivrud, I. M., Jore, S., Bakka, H., Vindenes, Y., & Mysterud, A. (2023). The emergence and shift in seasonality of Lyme borreliosis in Northern Europe. Proceedings of the Royal Society B: Biological Sciences, 290(1993), 20222420. https://doi.org/10.1098/rspb.2022.2420 Hoch, T., Madouasse, A., Jacquot, M., Wongnak, P., Beugnet, F., Bournez, L., . . . Agoulon, A. (2023). Seasonality of host-seeking Ixodes ricinus nymph abundance in relation to climate. bioRxiv, ver.4 peer-reviewed and recommended by Peer Community In Ecology. https://doi.org/10.1101/2022.07.25.501416 Uusitalo, R., Siljander, M., Lindén, A., Sormunen, J. J., Aalto, J., Hendrickx, G., . . . Vapalahti, O. (2022). Predicting habitat suitability for Ixodes ricinus and Ixodes persulcatus ticks in Finland. Parasites & Vectors, 15(1), 310. https://doi.org/10.1186/s13071-022-05410-8 Wongnak, P., Bord, S., Jacquot, M., Agoulon, A., Beugnet, F., Bournez, L., . . . Chalvet-Monfray, K. (2022). Meteorological and climatic variables predict the phenology of Ixodes ricinus nymph activity in France, accounting for habitat heterogeneity. Scientific Reports, 12(1), 7833. https://doi.org/10.1038/s41598-022-11479-z | Seasonality of host-seeking *Ixodes ricinus* nymph abundance in relation to climate | Thierry Hoch, Aurélien Madouasse, Maude Jacquot, Phrutsamon Wongnak, Fréderic Beugnet, Laure Bournez, Jean-François Cosson, Frédéric Huard, Sara Moutailler, Olivier Plantard, Valérie Poux, Magalie René-Martellet, Muriel Vayssier-Taussat, Hélène Ve... | <p style="text-align: justify;">There is growing concern about climate change and its impact on human health. Specifically, global warming could increase the probability of emerging infectious diseases, notably because of changes in the geographic... |  | Climate change, Population ecology, Statistical ecology | Nigel Yoccoz | 2022-10-14 18:43:56 | View | |
11 Mar 2024
Sex differences in the relationship between maternal and neonate cortisol in a free-ranging large mammalAmin, B., Fishman, R., Quinn, M., Matas, D., Palme, R., Koren, L., Ciuti, S. https://doi.org/10.1101/2023.05.04.538920Stress and stress hormones’ transmission from mothers to offspringRecommended by Matthieu Paquet based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Individuals can respond to environmental changes that they undergo directly (within-generation plasticity) but also through transgenerational plasticity, providing lasting effects that are transmitted to the next generations (Donelson et al. 2012; Munday et al. 2013; Kuijper & Hoyle 2015; Auge et al. 2017, Tariel et al. 2020). These parental effects can affect offspring via various mechanisms, notably via maternal transmission of hormones to the eggs or growing embryos (Mousseau & Fox 1998). While the effects of environmental quality may simply carry-over to the next generation (e.g., females in stressful environments give birth to offspring in poorer condition), parental effects may also be a mechanism that adjusts offspring phenotype in response to environmental variation and predictability, and thereby match offspring's phenotype to future environmental conditions (Gluckman et al. 2005; Marshall & Uller 2007; Dey et al. 2016; Yin et al. 2019), for example by preparing their offspring to an expected stressful environment. When females experience stress during gestation or egg formation, elevations in glucocorticoids (GC) are expected to affect offspring phenotype in many ways, from the offspring's own GC levels, to their growth and survival (Sheriff et al. 2017). This is a well established idea, but how strong is the evidence for this? A meta-analysis on birds found no clear effect of corticosterone manipulation on offspring traits (38 studies on 9 bird species for corticosterone manipulation; Podmokła et al. 2018). Another meta-analysis including 14 vertebrate species found no clear effect of prenatal stress on offspring GC (Thayer et al. 2018). Finally, a meta-analysis on wild vertebrates (23 species) found no clear effect of GC-mediated maternal effects on offspring traits (MacLeod et al. 2021). As often when facing such inconclusive results, context dependence has been suggested as one potential reason for such inconsistencies, for exemple sex specific effects (Groothuis et al. 2019, 2020). However, sex specific measures on offspring are scarce (Podmokła et al. 2018). Moreover, the literature available is still limited to a few, mostly “model” species. With their study, Amin et al. (2024) show the way to improve our understanding on GC transmission from mother to offspring and its effects in several aspects. First they used innovative non-invasive methods (which could broaden the range of species available to study) by quantifying cortisol metabolites from faecal samples collected from pregnant females, as proxy for maternal GC level, and relating it to GC levels from hairs of their neonate offspring. Second they used a free ranging large mammal (taxa from which literature is missing): the fallow deer (Dama dama). Third, they provide sex specific measures of GC levels. And finally but importantly, they are exemplary in their transparency regarding 1) the exploratory nature of their study, 2) their statistical thinking and procedure, and 3) the study limitations (e.g., low sample size and high within individual variation of measurements). I hope this study will motivate more research (on the fallow deer, and on other species) to broaden and strengthen our understanding of sex specific effects of maternal stress and CG levels on offspring phenotype and fitness. References Amin, B., Fishman, R., Quinn, M., Matas, D., Palme, R., Koren, L., & Ciuti, S. (2024). Sex differences in the relationship between maternal and foetal glucocorticoids in a free-ranging large mammal. bioRxiv, ver. 4 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.05.04.538920 Auge, G.A., Leverett, L.D., Edwards, B.R. & Donohue, K. (2017). Adjusting phenotypes via within-and across-generational plasticity. New Phytologist, 216, 343–349. https://doi.org/10.1111/nph.14495 Dey, S., Proulx, S.R. & Teotonio, H. (2016). Adaptation to temporally fluctuating environments by the evolution of maternal effects. PLoS biology, 14, e1002388. https://doi.org/10.1371/journal.pbio.1002388 Donelson, J.M., Munday, P.L., McCormick, M.I. & Pitcher, C.R. (2012). Rapid transgenerational acclimation of a tropical reef fish to climate change. Nature Climate Change, 2, 30. https://doi.org/10.1038/nclimate1323 Gluckman, P.D., Hanson, M.A. & Spencer, H.G. (2005). Predictive adaptive responses and human evolution. Trends in ecology & evolution, 20, 527–533. https://doi.org/10.1016/j.tree.2005.08.001 Groothuis, Ton GG, Bin-Yan Hsu, Neeraj Kumar, and Barbara Tschirren. "Revisiting mechanisms and functions of prenatal hormone-mediated maternal effects using avian species as a model." Philosophical Transactions of the Royal Society B 374, no. 1770 (2019): 20180115. https://doi.org/10.1098/rstb.2018.0115 Groothuis, Ton GG, Neeraj Kumar, and Bin-Yan Hsu. "Explaining discrepancies in the study of maternal effects: the role of context and embryo." Current Opinion in Behavioral Sciences 36 (2020): 185-192. https://doi.org/10.1016/j.cobeha.2020.10.006 Kuijper, B. & Hoyle, R.B. (2015). When to rely on maternal effects and when on phenotypic plasticity? Evolution, 69, 950–968. https://doi.org/10.1111/evo.12635 MacLeod, Kirsty J., Geoffrey M. While, and Tobias Uller. "Viviparous mothers impose stronger glucocorticoid‐mediated maternal stress effects on their offspring than oviparous mothers." Ecology and Evolution 11, no. 23 (2021): 17238-17259. Marshall, D.J. & Uller, T. (2007). When is a maternal effect adaptive? Oikos, 116, 1957–1963. https://doi.org/10.1111/j.2007.0030-1299.16203.x Mousseau, T.A. & Fox, C.W. (1998). Maternal effects as adaptations. Oxford University Press. Munday, P.L., Warner, R.R., Monro, K., Pandolfi, J.M. & Marshall, D.J. (2013). Predicting evolutionary responses to climate change in the sea. Ecology Letters, 16, 1488–1500. https://doi.org/10.1111/ele.12185 Podmokła, Edyta, Szymon M. Drobniak, and Joanna Rutkowska. "Chicken or egg? Outcomes of experimental manipulations of maternally transmitted hormones depend on administration method–a meta‐analysis." Biological Reviews 93, no. 3 (2018): 1499-1517. https://doi.org/10.1111/brv.12406 Sheriff, M. J., Bell, A., Boonstra, R., Dantzer, B., Lavergne, S. G., McGhee, K. E., MacLeod, K. J., Winandy, L., Zimmer, C., & Love, O. P. (2017). Integrating ecological and evolutionary context in the study of maternal stress. Integrative and Comparative Biology, 57(3), 437–449. https://doi.org/10.1093/icb/icx105 Tariel, Juliette, Sandrine Plénet, and Émilien Luquet. "Transgenerational plasticity in the context of predator-prey interactions." Frontiers in Ecology and Evolution 8 (2020): 548660. https://doi.org/10.3389/fevo.2020.548660 Thayer, Zaneta M., Meredith A. Wilson, Andrew W. Kim, and Adrian V. Jaeggi. "Impact of prenatal stress on offspring glucocorticoid levels: A phylogenetic meta-analysis across 14 vertebrate species." Scientific Reports 8, no. 1 (2018): 4942. https://doi.org/10.1038/s41598-018-23169-w Yin, J., Zhou, M., Lin, Z., Li, Q.Q. & Zhang, Y.-Y. (2019). Transgenerational effects benefit offspring across diverse environments: a meta-analysis in plants and animals. Ecology letters, 22, 1976–1986. https://doi.org/10.1111/ele.13373 | Sex differences in the relationship between maternal and neonate cortisol in a free-ranging large mammal | Amin, B., Fishman, R., Quinn, M., Matas, D., Palme, R., Koren, L., Ciuti, S. | <p style="text-align: justify;">Maternal phenotypes can have long-term effects on offspring phenotypes. These maternal effects may begin during gestation, when maternal glucocorticoid (GC) levels may affect foetal GC levels, thereby having an orga... | Evolutionary ecology, Maternal effects, Ontogeny, Physiology, Zoology | Matthieu Paquet | 2023-06-05 09:06:56 | View | ||
22 May 2019

Sex makes them sleepy: host reproductive status induces diapause in a parasitoid population experiencing harsh wintersTougeron K., Brodeur J., van Baaren J., Renault D. and Le Lann C. https://doi.org/10.1101/371385The response of interacting species to biotic seasonal cuesRecommended by Adele Mennerat and Enric Frago based on reviews by Anne Duplouy and 1 anonymous reviewerIn temperate regions, food abundance and quality vary greatly throughout the year, and the ability of organisms to synchronise their phenology to these changes is a key determinant of their reproductive success. Successful synchronisation requires that cues are perceived prior to change, leaving time for physiological adjustments. References [1] Tauber, M. J., Tauber, C. A., and Masaki, S. (1986). Seasonal Adaptations of Insects. Oxford, New York: Oxford University Press. | Sex makes them sleepy: host reproductive status induces diapause in a parasitoid population experiencing harsh winters | Tougeron K., Brodeur J., van Baaren J., Renault D. and Le Lann C. | <p>When organisms coevolve, any change in one species can induce phenotypic changes in traits and ecology of the other species. The role such interactions play in ecosystems is central, but their mechanistic bases remain underexplored. Upper troph... |  | Coexistence, Evolutionary ecology, Experimental ecology, Host-parasite interactions, Physiology | Adele Mennerat | 2018-07-18 18:51:03 | View | |
31 May 2022

Sexual coercion in a natural mandrill populationNikolaos Smit, Alice Baniel, Berta Roura-Torres, Paul Amblard-Rambert, Marie J. E. Charpentier, Elise Huchard https://doi.org/10.1101/2022.02.07.479393Rare behaviours can have strong effects: evidence for sexual coercion in mandrillsRecommended by Matthieu Paquet based on reviews by Micaela Szykman Gunther and 1 anonymous reviewer based on reviews by Micaela Szykman Gunther and 1 anonymous reviewer
Sexual coercion can be defined as the use by a male of force, or threat of force, which increases the chances that a female will mate with him at a time when she is likely to be fertile, and/or decrease the chances that she will mate with other males, at some cost to the female (Smuts & Smuts 1993). It has been evidenced in a wide range of species and may play an important role in the evolution of sexual conflict and social systems. However, identifying sexual coercion in natural systems can be particularly challenging. Notably, while male behaviour may have immediate consequences on mating success (“harassment”), the mating benefits may be delayed in time (“intimidation”), and in such cases, evidencing coercion requires detailed temporal data at the individual level. Moreover, in some species male aggressive behaviours may be subtle or rare and hence hardly observed, yet still have important effects on female mating probability and fitness. Therefore, investigating the occurrence and consequences of sexual coercion in such species is particularly relevant but studying it in a statistically robust way is likely to require a considerable amount of time spent observing individuals. In this paper, Smit et al. (2022) test three clear predictions of the sexual coercion hypothesis in a natural population of Mandrills, where severe male aggression towards females is rare: (1) male aggression is more likely on sexually receptive females than on females in other reproductive states, (2) receptive females are more likely to be injured and (3) male aggression directed towards females is positively related to subsequent probability of copulation between those dyads. They also tested an alternative hypothesis, the “aggressive male phenotype” under which the correlation between male aggression towards females and subsequent mating could be statistically explained by male overall aggressivity. In agreement with the three predictions of the sexual coercion hypothesis, (1) male aggression was on average 5 times more likely, and (2) injuries twice as likely, to be observed on sexually receptive females than on females in other reproductive states and (3) copulation between males and sexually receptive females was twice more likely to be observed when aggression by this male was observed on the female before sexual receptivity. There was no support for the aggressive male hypothesis. The reviewers and I were highly positive about this study, notably regarding the way it is written and how the predictions are carefully and clearly stated, tested, interpreted, and discussed. This study is a good illustration of a case where some behaviours may not be common or obvious yet have strong effects and likely important consequences and thus be clearly worth studying. More generally, it shows once more the importance of detailed long-term studies at the individual level for our understanding of the ecology and evolution of wild populations. It is also a good illustration of the challenges faced, when comparing the likelihood of contrasting hypotheses means we need to alter sample sizes and/or the likelihood to observe at all some behaviours. For example, observing copulation within minutes after aggression (and therefore, showing statistical support for “harassment”) is inevitably less likely than observing copulations on the longer-term (and therefore showing statistical support for “intimidation”, when of course effort is put into recording such behavioural data on the long-term). Such challenges might partly explain some apparently intriguing results. For example, why are swollen females more aggressed by males if only aggression before the swollen period seems associated with more chances of mating? Here, the authors systematically provide effect sizes (and confidence intervals) and often describe the effects in an intuitive biological way (e.g., “Swollen females were, on average, about five times more likely to become injured”). This clearly helps the reader to not merely compare statistical significances but also the biological strengths of the estimated effects and the uncertainty around them. They also clearly acknowledge limits due to sample size when testing the harassment hypothesis, yet they provide precious information on the probability of observing mating (a rare behaviour) directly after aggression (already a rare behaviour!), that is, 3 times out of 38 aggressions observed between a male and a swollen female. Once again, this highlights how important it is to be able to pursue the enormous effort put so far into closely and continuously monitoring this wild population. Finally, this study raises exciting new questions, notably regarding to what extent females exhibit “counter-strategies” in response to sexual coercion, notably whether there is still scope for female mate choice under such conditions, and what are the fitness consequences of these dynamic conflicting sexual interactions. No doubt these questions will sooner than later be addressed by the authors, and I am looking forward to reading their upcoming work. References Smit N, Baniel A, Roura-Torres B, Amblard-Rambert P, Charpentier MJE, Huchard E (2022) Sexual coercion in a natural mandrill population. bioRxiv, 2022.02.07.479393, ver. 5 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2022.02.07.479393 Smuts BB, Smuts R w. (1993) Male Aggression and Sexual Coercion of Females in Nonhuman Primates and Other Mammals: Evidence and Theoretical Implications. In: Advances in the Study of Behavior (eds Slater PJB, Rosenblatt JS, Snowdon CT, Milinski M), pp. 1–63. Academic Press. https://doi.org/10.1016/S0065-3454(08)60404-0 | Sexual coercion in a natural mandrill population | Nikolaos Smit, Alice Baniel, Berta Roura-Torres, Paul Amblard-Rambert, Marie J. E. Charpentier, Elise Huchard | <p style="text-align: justify;">Increasing evidence indicates that sexual coercion is widespread. While some coercive strategies are conspicuous, such as forced copulation or sexual harassment, less is known about the ecology and evolution of inti... |  | Behaviour & Ethology | Matthieu Paquet | 2022-02-11 09:32:49 | View | |
20 Jun 2019

Sexual segregation in a highly pagophilic and sexually dimorphic marine predatorChristophe Barbraud, Karine Delord, Akiko Kato, Paco Bustamante, Yves Cherel https://doi.org/10.1101/472431Sexual segregation in a sexually dimorphic seabird: a matter of spatial scaleRecommended by Denis Réale based on reviews by Dries Bonte and 1 anonymous reviewerSexual segregation appears in many taxa and can have important ecological, evolutionary and conservation implications. Sexual segregation can take two forms: either the two sexes specialise in different habitats but share the same area (habitat segregation), or they occupy the same habitat but form separate, unisex groups (social segregation) [1,2]. Segregation would have evolved as a way to avoid, or at least, reduce intersexual competition. References [1] Conradt, L. (2005). Definitions, hypotheses, models and measures in the study of animal segregation. In Sexual segregation in vertebrates: ecology of the two sexes (Ruckstuhl K.E. and Neuhaus, P. eds). Cambridge University Press, Cambridge, United Kingdom. Pp:11–34. | Sexual segregation in a highly pagophilic and sexually dimorphic marine predator | Christophe Barbraud, Karine Delord, Akiko Kato, Paco Bustamante, Yves Cherel | <p>Sexual segregation is common in many species and has been attributed to intra-specific competition, sex-specific differences in foraging efficiency or in activity budgets and habitat choice. However, very few studies have simultaneously quantif... |  | Foraging, Marine ecology | Denis Réale | Dries Bonte, Anonymous | 2018-11-19 13:40:59 | View |
11 Mar 2021
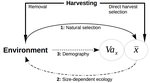
Size-dependent eco-evolutionary feedbacks in fisheriesEric Edeline and Nicolas Loeuille https://doi.org/10.1101/2020.04.03.022905“Hidden” natural selection and the evolution of body size in harvested stocksRecommended by Simon Blanchet based on reviews by Jean-François Arnoldi and 1 anonymous reviewerHumans are exploiting biological resources since thousands of years. Exploitation of biological resources has become particularly intense since the beginning of the 20th century and the steep increase in the worldwide human population size. Marine and freshwater fishes are not exception to that rule, and they have been (and continue to be) strongly harvested as a source of proteins for humans. For some species, fishery has been so intense that natural stocks have virtually collapsed in only a few decades. The worst example begin that of the Northwest Atlantic cod that has declined by more than 95% of its historical biomasses in only 20-30 years of intensive exploitation (Frank et al. 2005). These rapid and steep changes in biomasses have huge impacts on the entire ecosystems since species targeted by fisheries are often at the top of trophic chains (Frank et al. 2005). Beyond demographic impacts, fisheries also have evolutionary impacts on populations, which can also indirectly alter ecosystems (Uusi-Heikkilä et al. 2015; Palkovacs et al. 2018). Fishermen generally focus on the largest specimens, and hence exert a strong selective pressure against these largest fish (which is called “harvest selection”). There is now ample evidence that harvest selection can lead to rapid evolutionary changes in natural populations toward small individuals (Kuparinen & Festa-Bianchet 2017). These evolutionary changes are of course undesirable from a human perspective, and have attracted many scientific questions. Nonetheless, the consequence of harvest selection is not always observable in natural populations, and there are cases in which no phenotypic change (or on the contrary an increase in mean body size) has been observed after intense harvest pressures. In a conceptual Essay, Edeline and Loeuille (Edeline & Loeuille 2020) propose novel ideas to explain why the evolutionary consequences of harvest selection can be so diverse, and how a cross talk between ecological and evolutionary dynamics can explain patterns observed in natural stocks. The general and novel concept proposed by Edeline and Loeuille is actually as old as Darwin’s book; The Origin of Species (Darwin 1859). It is based on the simple idea that natural selection acting on harvested populations can actually be strong, and counter-balance (or on the contrary reinforce) the evolutionary consequence of harvest selection. Although simple, the idea that natural and harvest selection are jointly shaping contemporary evolution of exploited populations lead to various and sometimes complex scenarios that can (i) explain unresolved empirical patterns and (ii) refine predictions regarding the long-term viability of exploited populations. The Edeline and Loeuille’s crafty inspiration is that natural selection acting on exploited populations is itself an indirect consequence of harvest (Edeline & Loeuille 2020). They suggest that, by modifying the size structure of populations (a key parameter for ecological interactions), harvest indirectly alters interactions between populations and their biotic environment through competition and predation, which changes the ecological theatre and hence the selective pressures acting back to populations. They named this process “size-dependent eco-evolutionary feedback loops” and develop several scenarios in which these feedback loops ultimately deviate the evolutionary outcome of harvest selection from expectation. The scenarios they explore are based on strong theoretical knowledge, and range from simple ones in which a single species (the harvest species) is evolving to more complex (and realistic) ones in which multiple (e.g. the harvest species and its prey) species are co-evolving. I will not come into the details of each scenario here, and I will let the readers (re-)discovering the complex beauty of biological life and natural selection. Nonetheless, I will emphasize the importance of considering these eco-evolutionary processes altogether to fully grasp the response of exploited populations. Edeline and Loeuille convincingly demonstrate that reduced body size due to harvest selection is obviously not the only response of exploited fish populations when natural selection is jointly considered (Edeline & Loeuille 2020). On the contrary, they show that –under some realistic ecological circumstances relaxing exploitative competition due to reduced population densities- natural selection can act antagonistically, and hence favour stable body size in exploited populations. Although this seems further desirable from a human perspective than a downsizing of exploited populations, it is actually mere window dressing as Edeline and Loeuille further showed that this response is accompanied by an erosion of the evolvability –and hence a lowest probability of long-term persistence- of these exploited populations. Humans, by exploiting biological resources, are breaking the relative equilibrium of complex entities, and the response of populations to this disturbance is itself often complex and heterogeneous. In this Essay, Edeline and Loeuille provide –under simple terms- the theoretical and conceptual bases required to improve predictions regarding the evolutionary responses of natural populations to exploitation by humans (Edeline & Loeuille 2020). An important next step will be to generate data and methods allowing confronting the empirical reality to these novel concepts (e.g. (Monk et al. 2021), so as to identify the most likely evolutionary scenarios sustaining biological responses of exploited populations, and hence to set the best management plans for the long-term sustainability of these populations. References Darwin, C. (1859). On the Origin of Species by Means of Natural Selection. John Murray, London. Edeline, E. & Loeuille, N. (2021) Size-dependent eco-evolutionary feedbacks in fisheries. bioRxiv, 2020.04.03.022905, ver. 4 peer-reviewed and recommended by PCI Ecology. doi: https://doi.org/10.1101/2020.04.03.022905 Frank, K.T., Petrie, B., Choi, J. S. & Leggett, W.C. (2005). Trophic Cascades in a Formerly Cod-Dominated Ecosystem. Science, 308, 1621–1623. doi: https://doi.org/10.1126/science.1113075 Kuparinen, A. & Festa-Bianchet, M. (2017). Harvest-induced evolution: insights from aquatic and terrestrial systems. Philos. Trans. R. Soc. B Biol. Sci., 372, 20160036. doi: https://doi.org/10.1098/rstb.2016.0036 Monk, C.T., Bekkevold, D., Klefoth, T., Pagel, T., Palmer, M. & Arlinghaus, R. (2021). The battle between harvest and natural selection creates small and shy fish. Proc. Natl. Acad. Sci., 118, e2009451118. doi: https://doi.org/10.1073/pnas.2009451118 Palkovacs, E.P., Moritsch, M.M., Contolini, G.M. & Pelletier, F. (2018). Ecology of harvest-driven trait changes and implications for ecosystem management. Front. Ecol. Environ., 16, 20–28. doi: https://doi.org/10.1002/fee.1743 Uusi-Heikkilä, S., Whiteley, A.R., Kuparinen, A., Matsumura, S., Venturelli, P.A., Wolter, C., et al. (2015). The evolutionary legacy of size-selective harvesting extends from genes to populations. Evol. Appl., 8, 597–620. doi: https://doi.org/10.1111/eva.12268 | Size-dependent eco-evolutionary feedbacks in fisheries | Eric Edeline and Nicolas Loeuille | <p>Harvesting may drive body downsizing along with population declines and decreased harvesting yields. These changes are commonly construed as direct consequences of harvest selection, where small-bodied, early-reproducing individuals are immedia... |  | Biodiversity, Community ecology, Competition, Eco-evolutionary dynamics, Evolutionary ecology, Food webs, Interaction networks, Life history, Population ecology, Theoretical ecology | Simon Blanchet | 2020-04-03 16:14:05 | View | |
19 Feb 2020

Soil variation response is mediated by growth trajectories rather than functional traits in a widespread pioneer Neotropical treeSébastien Levionnois, Niklas Tysklind, Eric Nicolini, Bruno Ferry, Valérie Troispoux, Gilles Le Moguedec, Hélène Morel, Clément Stahl, Sabrina Coste, Henri Caron, Patrick Heuret https://doi.org/10.1101/351197Growth trajectories, better than organ-level functional traits, reveal intraspecific response to environmental variationRecommended by François Munoz based on reviews by Georges Kunstler and François Munoz based on reviews by Georges Kunstler and François Munoz
Functional traits are “morpho-physio-phenological traits which impact fitness indirectly via their effects on growth, reproduction and survival” [1]. Most functional traits are defined at organ level, e.g. for leaves, roots and stems, and reflect key aspects of resource acquisition and resource use by organisms for their development and reproduction [2]. More rarely, some functional traits can be related to spatial development, such as vegetative height and lateral spread in plants. References [1] Violle, C., Navas, M. L., Vile, D., Kazakou, E., Fortunel, C., Hummel, I., & Garnier, E. (2007). Let the concept of trait be functional!. Oikos, 116(5), 882-892. doi: 10.1111/j.0030-1299.2007.15559.x | Soil variation response is mediated by growth trajectories rather than functional traits in a widespread pioneer Neotropical tree | Sébastien Levionnois, Niklas Tysklind, Eric Nicolini, Bruno Ferry, Valérie Troispoux, Gilles Le Moguedec, Hélène Morel, Clément Stahl, Sabrina Coste, Henri Caron, Patrick Heuret | <p style="text-align: justify;">1- Trait-environment relationships have been described at the community level across tree species. However, whether interspecific trait-environment relationships are consistent at the intraspecific level is yet unkn... |  | Botany, Eco-evolutionary dynamics, Habitat selection, Ontogeny, Tropical ecology | François Munoz | 2018-06-21 17:13:17 | View | |
15 Feb 2024
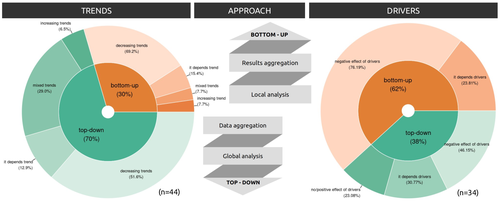
Sources of confusion in global biodiversity trendsMaelys Boennec, Vasilis Dakos, Vincent Devictor https://doi.org/10.32942/X29W3HUnraveling the Complexity of Global Biodiversity Dynamics: Insights and ImperativesRecommended by Paulo Borges based on reviews by Pedro Cardoso and 1 anonymous reviewer based on reviews by Pedro Cardoso and 1 anonymous reviewer
Biodiversity loss is occurring at an alarming rate across terrestrial and marine ecosystems, driven by various processes that degrade habitats and threaten species with extinction. Despite the urgency of this issue, empirical studies present a mixed picture, with some indicating declining trends while others show more complex patterns. In a recent effort to better understand global biodiversity dynamics, Boennec et al. (2024) conducted a comprehensive literature review examining temporal trends in biodiversity. Their analysis reveals that reviews and meta-analyses, coupled with the use of global indicators, tend to report declining trends more frequently. Additionally, the study underscores a critical gap in research: the scarcity of investigations into the combined impact of multiple pressures on biodiversity at a global scale. This lack of understanding complicates efforts to identify the root causes of biodiversity changes and develop effective conservation strategies. This study serves as a crucial reminder of the pressing need for long-term biodiversity monitoring and large-scale conservation studies. By filling these gaps in knowledge, researchers can provide policymakers and conservation practitioners with the insights necessary to mitigate biodiversity loss and safeguard ecosystems for future generations. References Boennec, M., Dakos, V. & Devictor, V. (2023). Sources of confusion in global biodiversity trend. bioRxiv, ver. 4 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.32942/X29W3H
| Sources of confusion in global biodiversity trends | Maelys Boennec, Vasilis Dakos, Vincent Devictor | <p>Populations and ecological communities are changing worldwide, and empirical studies exhibit a mixture of either declining or mixed trends. Confusion in global biodiversity trends thus remains while assessing such changes is of major social, po... |  | Biodiversity, Conservation biology, Meta-analyses | Paulo Borges | 2023-09-20 11:10:25 | View | |
26 May 2021

Spatial distribution of local patch extinctions drives recovery dynamics in metacommunitiesCamille Saade, Sonia Kéfi, Claire Gougat-Barbera, Benjamin Rosenbaum, and Emanuel A. Fronhofer https://doi.org/10.1101/2020.12.03.409524Unity makes strength: clustered extinctions have stronger, longer-lasting effects on metacommunities dynamicsRecommended by Elodie Vercken based on reviews by David Murray-Stoker and Frederik De LaenderIn this article, Saade et al. (2021) investigate how the rate of local extinctions and their spatial distribution affect recolonization dynamics in metacommunities. They use an elegant combination of microcosm experiments with metacommunities of freshwater ciliates and mathematical modelling mirroring their experimental system. Their main findings are (i) that local patch extinctions increase both local (α-) and inter-patch (β-) diversity in a transient way during the recolonization process, (ii) that these effects depend more on the spatial distribution of extinctions (dispersed or clustered) than on their amount, and (iii) that they may spread regionally. A major strength of this study is that it highlights the importance of considering the spatial structure explicitly. Recent work on ecological networks has shown repeatedly that network structure affects the propagation of pathogens (Badham and Stocker 2010), invaders (Morel-Journel et al. 2019), or perturbation events (Gilarranz et al. 2017). Here, the spatial structure of the metacommunity is a regular grid of patches, but the distribution of extinction events may be either regularly dispersed (i.e., extinct patches are distributed evenly over the grid and are all surrounded by non-extinct patches only) or clustered (all extinct patches are neighbours). This has a direct effect on the neighbourhood of perturbed patches, and because perturbations have mostly local effects, their recovery dynamics are dominated by the composition of this immediate neighbourhood. In landscapes with dispersed extinctions, the neighbourhood of a perturbed patch is not affected by the amount of extinctions, and neither is its recovery time. In contrast, in landscapes with clustered extinctions, the amount of extinctions affects the depth of the perturbed area, which takes longer to recover when it is larger. Interestingly, the spatial distribution of extinctions here is functionally equivalent to differences in connectivity between perturbed and unperturbed patches, which results in contrasted “rescue recovery” and “mixing recovery” regimes as described by Zelnick et al. (2019).
Levins R (1969) Some Demographic and Genetic Consequences of Environmental Heterogeneity for Biological Control1. Bulletin of the Entomological Society of America, 15, 237–240. https://doi.org/10.1093/besa/15.3.237 Ruokolainen L (2013) Spatio-Temporal Environmental Correlation and Population Variability in Simple Metacommunities. PLOS ONE, 8, e72325. https://doi.org/10.1371/journal.pone.0072325 Saade C, Kefi S, Gougat-Barbera C, Rosenbaum B, Fronhofer EA (2021) Spatial distribution of local patch extinctions drives recovery dynamics in metacommunities. bioRxiv, 2020.12.03.409524, ver. 4 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2020.12.03.409524 | Spatial distribution of local patch extinctions drives recovery dynamics in metacommunities | Camille Saade, Sonia Kéfi, Claire Gougat-Barbera, Benjamin Rosenbaum, and Emanuel A. Fronhofer | <p style="text-align: justify;">Human activities lead more and more to the disturbance of plant and animal communities with local extinctions as a consequence. While these negative effects are clearly visible at a local scale, it is less clear how... |  | Biodiversity, Coexistence, Colonization, Community ecology, Competition, Dispersal & Migration, Experimental ecology, Landscape ecology, Spatial ecology, Metacommunities & Metapopulations | Elodie Vercken | 2020-12-08 15:55:20 | View | |
27 Jan 2023
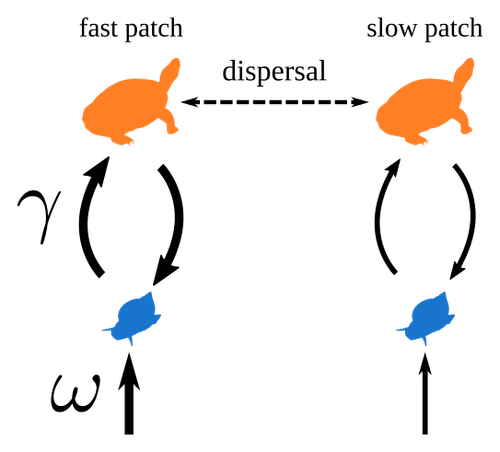
Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunitiesPierre Quévreux, Bart Haegeman and Michel Loreau https://hal.science/hal-03829838How does spatial heterogeneity affect stability of trophic metacommunities?Recommended by Werner Ulrich based on reviews by Phillip P.A. Staniczenko, Ludek Berec and Diogo Provete based on reviews by Phillip P.A. Staniczenko, Ludek Berec and Diogo Provete
The temporal or spatial variability in species population sizes and interaction strength of animal and plant communities has a strong impact on aggregate community properties (for instance biomass), community composition, and species richness (Kokkoris et al. 2002). Early work on spatial and temporal variability strongly indicated that asynchronous population and environmental fluctuations tend to stabilise community structures and diversity (e.g. Holt 1984, Tilman and Pacala 1993, McCann et al. 1998, Amarasekare and Nisbet 2001). Similarly, trophic networks might be stabilised by spatial heterogeneity (Hastings 1977) and an asymmetry of energy flows along food chains (Rooney et al. 2006). The interplay between temporal, spatial, and trophic heterogeneity within the meta-community concept has got much less interest. In the recent preprint in PCI Ecology, Quévreux et al. (2023) report that Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunities. These authors rightly notice that the interplay between trophic and spatial heterogeneity might induce contrasting effects depending on the internal dynamics of the system. Their contribution builds on prior work (Quévreux et al. 2021a, b) on perturbed trophic cascades. I found this paper particularly interesting because it is in the, now century-old, tradition to show that ecological things are not so easy. Since the 1930th, when Nicholson and Baily and others demonstrated that simple deterministic population models might generate stability and (pseudo-)chaos ecologists have realised that systems triggered by two or more independent processes might be intrinsically unpredictable and generate different outputs depending on the initial parameter settings. This resembles the three-body problem in physics. The present contribution of Quévreux et al. (2023) extends this knowledge to an example of a spatially explicit trophic model. Their main take-home message is that asymmetric energy flows in predator–prey relationships might have contrasting effects on the stability of metacommunities receiving localised perturbations. Stability is context dependent. Of course, the work is merely a theoretical exercise using a simplistic trophic model. It demands verification with field data. Nevertheless, we might expect even stronger unpredictability in more realistic multitrophic situations. Therefore, it should be seen as a proof of concept. Remember that increasing trophic connectance tends to destabilise food webs (May 1972). In this respect, I found the final outlook to bioconservation ambitious but substantiated. Biodiversity management needs a holistic approach focusing on all aspects of ecological functioning. I would add the need to see stability and biodiversity within an evolutionary perspective. References Amarasekare P, Nisbet RM (2001) Spatial Heterogeneity, Source‐Sink Dynamics, and the Local Coexistence of Competing Species. The American Naturalist, 158, 572–584. https://doi.org/10.1086/323586 Hastings A (1977) Spatial heterogeneity and the stability of predator-prey systems. Theoretical Population Biology, 12, 37–48. https://doi.org/10.1016/0040-5809(77)90034-X Holt RD (1984) Spatial Heterogeneity, Indirect Interactions, and the Coexistence of Prey Species. The American Naturalist, 124, 377–406. https://doi.org/10.1086/284280 Kokkoris GD, Jansen VAA, Loreau M, Troumbis AY (2002) Variability in interaction strength and implications for biodiversity. Journal of Animal Ecology, 71, 362–371. https://doi.org/10.1046/j.1365-2656.2002.00604.x May RM (1972) Will a Large Complex System be Stable? Nature, 238, 413–414. https://doi.org/10.1038/238413a0 McCann K, Hastings A, Huxel GR (1998) Weak trophic interactions and the balance of nature. Nature, 395, 794–798. https://doi.org/10.1038/27427 Quévreux P, Barbier M, Loreau M (2021) Synchrony and Perturbation Transmission in Trophic Metacommunities. The American Naturalist, 197, E188–E203. https://doi.org/10.1086/714131 Quévreux P, Pigeault R, Loreau M (2021) Predator avoidance and foraging for food shape synchrony and response to perturbations in trophic metacommunities. Journal of Theoretical Biology, 528, 110836. https://doi.org/10.1016/j.jtbi.2021.110836 Quévreux P, Haegeman B, Loreau M (2023) Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunities. hal-03829838, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://hal.science/hal-03829838 Rooney N, McCann K, Gellner G, Moore JC (2006) Structural asymmetry and the stability of diverse food webs. Nature, 442, 265–269. https://doi.org/10.1038/nature04887 Tilman D, Pacala S (1993) The maintenance of species richness in plant communities. In: Ricklefs, R.E., Schluter, D. (eds) Species Diversity in Ecological Communities: Historical and Geographical Perspectives. University of Chicago Press, pp. 13–25. | Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunities | Pierre Quévreux, Bart Haegeman and Michel Loreau | <p> Spatial heterogeneity is a fundamental feature of ecosystems, and ecologists have identified it as a factor promoting the stability of population dynamics. In particular, differences in interaction strengths and resource supply between pa... |  | Dispersal & Migration, Food webs, Interaction networks, Spatial ecology, Metacommunities & Metapopulations, Theoretical ecology | Werner Ulrich | 2022-10-26 13:38:34 | View |
MANAGING BOARD
Julia Astegiano
Tim Coulson
Anna Eklof
Dominique Gravel
François Massol
Ben Phillips
Cyrille Violle










