Direct submissions to PCI Ecology from bioRxiv.org are possible using the B2J service
Latest recommendations

| Id | Title * | Authors * | Abstract * | Picture * | Thematic fields * | Recommender▲ | Reviewers | Submission date | |
|---|---|---|---|---|---|---|---|---|---|
16 Jun 2020
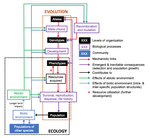
Environmental perturbations and transitions between ecological and evolutionary equilibria: an eco-evolutionary feedback frameworkTim Coulson https://doi.org/10.1101/509067Stasis and the phenotypic gambitRecommended by Tom Van Dooren based on reviews by Jacob Johansson, Katja Räsänen and 1 anonymous reviewerThe preprint "Environmental perturbations and transitions between ecological and evolutionary equilibria: an eco-evolutionary feedback framework" by Coulson (2020) presents a general framework for evolutionary ecology, useful to interpret patterns of selection and evolutionary responses to environmental transitions. The paper is written in an accessible and intuitive manner. It reviews important concepts which are at the heart of evolutionary ecology. Together, they serve as a worldview which you can carry with you to interpret patterns in data or observations in nature. I very much appreciate it that Coulson (2020) presents his personal intuition laid bare, the framework he uses for his research and how several strong concepts from theoretical ecology fit in there. Overviews as presented in this paper are important to understand how we as researchers put the pieces together. References [1] Coulson, T. (2020) Environmental perturbations and transitions between ecological and evolutionary equilibria: an eco-evolutionary feedback framework. bioRxiv, 509067, ver. 4 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/509067 | Environmental perturbations and transitions between ecological and evolutionary equilibria: an eco-evolutionary feedback framework | Tim Coulson | <p>I provide a general framework for linking ecology and evolution. I start from the fact that individuals require energy, trace molecules, water, and mates to survive and reproduce, and that phenotypic resource accrual traits determine an individ... |  | Eco-evolutionary dynamics, Evolutionary ecology | Tom Van Dooren | 2019-01-03 10:05:16 | View | |
13 May 2023

Symbiotic nutrient cycling enables the long-term survival of Aiptasia in the absence of heterotrophic food sourcesNils Radecker, Anders Meibom https://doi.org/10.1101/2022.12.07.519152Constraining the importance of heterotrophic vs autotrophic feeding in photosymbiotic cnidariansRecommended by Ulisse Cardini based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
The symbiosis with autotrophic dinoflagellate algae has enabled heterotrophic Cnidaria to thrive in nutrient-poor tropical waters (Muscatine and Porter 1977; Stanley 2006). In particular, mixotrophy, i.e. the ability to acquire nutrients through both autotrophy and heterotrophy, confers a competitive edge in oligotrophic waters, allowing photosymbiotic Cnidaria to outcompete benthic organisms limited to a single diet (e.g., McCook 2001). However, the relative importance of autotrophy vs heterotrophy in sustaining symbiotic cnidarian’s nutrition is still the subject of intense research. In fact, figuring out the cellular mechanisms by which symbiotic Cnidaria acquire a balanced diet for their metabolism and growth is relevant to our understanding of their physiology under varying environmental conditions and in response to anthropogenic perturbations. In this study's long-term starvation experiment, Radecker & Meibom (2023) investigated the survival of the photosymbiotic sea anemone Aiptasia in the absence of heterotrophic feeding. After one year of heterotrophic starvation, Apitasia anemones remained fully viable but showed an 85 % reduction in biomass. Using 13C-bicarbonate and 15N-ammonium labeling, electron microscopy and NanoSIMS imaging, the authors could clearly show that the contribution of algal-derived nutrients to the host metabolism remained unaffected as a result of increased algal photosynthesis and more efficient carbon translocation. At the same time, the absence of heterotrophic feeding caused severe nitrogen limitation in the starved Apitasia anemones. Overall, this study provides valuable insights into nutrient exchange within the symbiosis between Cnidaria and dinoflagellate algae at the cellular level and sheds new light on the importance of heterotrophic feeding as a nitrogen acquisition strategy for holobiont growth in oligotrophic waters. REFERENCES McCook L (2001) Competition between corals and algal turfs along a gradient of terrestrial influence in the nearshore central Great Barrier Reef. Coral Reefs 19:419–425. https://doi.org/10.1007/s003380000119 Muscatine L, Porter JW (1977) Reef corals: mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454–460. https://doi.org/10.2307/1297526 Radecker N, Meibom A (2023) Symbiotic nutrient cycling enables the long-term survival of Aiptasia in the absence of heterotrophic food sources. bioRxiv, ver. 3 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2022.12.07.519152 Stanley GD Jr (2006) Photosymbiosis and the evolution of modern coral reefs. Science 312:857–858. https://doi.org/10.1126/science.1123701 | Symbiotic nutrient cycling enables the long-term survival of Aiptasia in the absence of heterotrophic food sources | Nils Radecker, Anders Meibom | <p style="text-align: justify;">Phototrophic Cnidaria are mixotrophic organisms that can complement their heterotrophic diet with nutrients assimilated by their algal endosymbionts. Metabolic models suggest that the translocation of photosynthates... |  | Eco-evolutionary dynamics, Microbial ecology & microbiology, Symbiosis | Ulisse Cardini | 2022-12-12 10:50:55 | View | |
11 Oct 2023

Identification of microbial exopolymer producers in sandy and muddy intertidal sediments by compound-specific isotope analysisCédric Hubas, Julie Gaubert-Boussarie, An-Sofie D’Hondt, Bruno Jesus, Dominique Lamy, Vona Meleder, Antoine Prins, Philippe Rosa, Willem Stock, Koen Sabbe https://doi.org/10.1101/2022.12.02.516908Disentangling microbial exopolymer dynamics in intertidal sedimentsRecommended by Ute Risse-Buhl and Nils Rädecker based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
The secretion of extracellular polymeric substances (EPS) enables microorganisms to shape and interact with their environment [1]. EPS support cell adhesion and motility, offer protection from unfavorable conditions, and facilitate nutrient acquisition and transfer between microorganisms [2]. EPS production and consumption thus control the formation and structural organization of biofilms [3]. However, in marine environments, our understanding of the sources and composition of EPS is limited. References
| Identification of microbial exopolymer producers in sandy and muddy intertidal sediments by compound-specific isotope analysis | Cédric Hubas, Julie Gaubert-Boussarie, An-Sofie D’Hondt, Bruno Jesus, Dominique Lamy, Vona Meleder, Antoine Prins, Philippe Rosa, Willem Stock, Koen Sabbe | <p style="text-align: justify;">Extracellular polymeric substances (EPS) refer to a wide variety of high molecular weight molecules secreted outside the cell membrane by biofilm microorganisms. In the present study, EPS from marine microphytobenth... |  | Biodiversity, Ecological stoichiometry, Ecosystem functioning, Food webs, Marine ecology, Microbial ecology & microbiology, Soil ecology | Ute Risse-Buhl | 2022-12-06 14:13:11 | View | |
19 Dec 2020
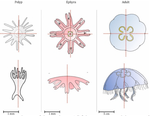
Hough transform implementation to evaluate the morphological variability of the moon jellyfish (Aurelia spp.)Céline Lacaux, Agnès Desolneux, Justine Gadreaud, Bertrand Martin-Garin and Alain Thiéry https://doi.org/10.1101/2020.03.11.986984A new member of the morphometrics jungle to better monitor vulnerable lagoonsRecommended by Vincent Bonhomme based on reviews by Julien Claude and 1 anonymous reviewerIn the recent years, morphometrics, the quantitative description of shape and its covariation [1] gained considerable momentum in evolutionary ecology. Using the form of organisms to describe, classify and try to understand their diversity can be traced back at least to Aristotle. More recently, two successive revolutions rejuvenated this idea [1–3]: first, a proper mathematical refoundation of the theory of shape, then a technical revolution in the apparatus able to acquire raw data. By using a feature extraction method and planning its massive use on data acquired by aerial drones, the study by Lacaux and colleagues [4] retraces this curse of events. The sample sizes studied here were too low to allow finer-grained ecophysiological investigations. That being said, the proof-of-concept is convincing and this paper paths the way for an operational and innovative approach to the ecological monitoring of sensible aquatic ecosystems. References [1] Kendall, D. G. (1989). A survey of the statistical theory of shape. Statistical Science, 87-99. doi: https://doi.org/10.1214/ss/1177012589 | Hough transform implementation to evaluate the morphological variability of the moon jellyfish (Aurelia spp.) | Céline Lacaux, Agnès Desolneux, Justine Gadreaud, Bertrand Martin-Garin and Alain Thiéry | <p>Variations of the animal body plan morphology and morphometry can be used as prognostic tools of their habitat quality. The potential of the moon jellyfish (Aurelia spp.) as a new model organism has been poorly tested. However, as a tetramerous... |  | Morphometrics | Vincent Bonhomme | 2020-03-18 17:40:51 | View | |
28 Mar 2019

Direct and transgenerational effects of an experimental heat wave on early life stages in a freshwater snailKatja Leicht, Otto Seppälä https://doi.org/10.1101/449777Escargots cooked just right: telling apart the direct and indirect effects of heat waves in freashwater snailsRecommended by vincent calcagno based on reviews by Amanda Lynn Caskenette, Kévin Tougeron and arnaud sentisAmongst the many challenges and forms of environmental change that organisms face in our era of global change, climate change is perhaps one of the most straightforward and amenable to investigation. First, measurements of day-to-day temperatures are relatively feasible and accessible, and predictions regarding the expected trends in Earth surface temperature are probably some of the most reliable we have. It appears quite clear, in particular, that beyond the overall increase in average temperature, the heat waves locally experienced by organisms in their natural habitats are bound to become more frequent, more intense, and more long-lasting [1]. Second, it is well appreciated that temperature is a major environmental factor with strong impacts on different facets of organismal development and life-history [2-4]. These impacts have reasonably clear mechanistic underpinnings, with definite connections to biochemistry, physiology, and considerations on energetics. Third, since variation in temperature is a challenge already experienced by natural populations across their current and historical ranges, it is not a completely alien form of environmental change. Therefore, we already learnt quite a lot about it in several species, and so did the species, as they may be expected to have evolved dedicated adaptive mechanisms to respond to elevated temperatures. Last, but not least, temperature is quite amenable to being manipulated as an experimental factor. References [1] Meehl, G. A., & Tebaldi, C. (2004). More intense, more frequent, and longer lasting heat waves in the 21st century. Science (New York, N.Y.), 305(5686), 994–997. doi: 10.1126/science.1098704 | Direct and transgenerational effects of an experimental heat wave on early life stages in a freshwater snail | Katja Leicht, Otto Seppälä | <p>Global climate change imposes a serious threat to natural populations of many species. Estimates of the effects of climate change‐mediated environmental stresses are, however, often based only on their direct effects on organisms, and neglect t... |  | Climate change | vincent calcagno | 2018-10-22 22:19:22 | View | |
06 Sep 2019
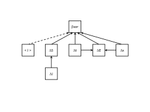
Assessing metacommunity processes through signatures in spatiotemporal turnover of community compositionFranck Jabot, Fabien Laroche, Francois Massol, Florent Arthaud, Julie Crabot, Maxime Dubart, Simon Blanchet, Francois Munoz, Patrice David, Thibault Datry https://doi.org/10.1101/480335On the importance of temporal meta-community dynamics for our understanding of assembly processesRecommended by Werner Ulrich based on reviews by Joaquín Hortal and 2 anonymous reviewers based on reviews by Joaquín Hortal and 2 anonymous reviewers
The processes that trigger community assembly are still in the centre of ecological interest. While prior work mostly focused on spatial patterns of co-occurrence within a meta-community framework [reviewed in 1, 2] recent studies also include temporal patterns of community composition [e.g. 3, 4, 5, 6]. In this preprint [7], Franck Jabot and co-workers extend they prior approaches to quasi neutral community assembly [8, 9, 10] and develop an analytical framework of spatial and temporal diversity turnover. A simple and heuristic path model for beta diversity and an extended ecological drift model serve as starting points. The model can be seen as a counterpart to Ulrich et al. [5]. These authors implemented competitive hierarchies into their neutral meta-community model while the present paper focuses on environmental filtering. Most important, the model and parameterization of four empirical data sets on aquatic plant and animal meta-communities used by Jabot et al. returned a consistent high influence of environmental stochasticity on species turnover. Of course, this major result does not come to a surprise. As typical for this kind of models it depends also to a good deal on the initial model settings. It nevertheless makes a strong conceptual point for the importance of environmental variability over dispersal and richness effects. One interesting side effect regards the impact of richness differences (ΔS). Jabot et al. interpret this as a ‘nuisance variable’ as they do not have a stringent explanation. Of course, it might be a pure statistical bias introduced by the Soerensen metric of turnover that is normalized by richness. However, I suspect that there is more behind the ΔS effect. Richness differences are generally associated with respective differences in total abundances and introduce source – sink dynamics that inevitably shape subsequent colonization – extinction processes. It would be interesting to see whether ΔS alone is able to trigger observed patterns of community assembly and community composition. Such an analysis would require partitioning of species turnover into richness and nestedness effects [11]. I encourage Jabot et al. to undertake such an effort. References [1] Götzenberger, L. et al. (2012). Ecological assembly rules in plant communities—approaches, patterns and prospects. Biological reviews, 87(1), 111-127. doi: 10.1111/j.1469-185X.2011.00187.x | Assessing metacommunity processes through signatures in spatiotemporal turnover of community composition | Franck Jabot, Fabien Laroche, Francois Massol, Florent Arthaud, Julie Crabot, Maxime Dubart, Simon Blanchet, Francois Munoz, Patrice David, Thibault Datry | <p>Although metacommunity ecology has been a major field of research in the last decades, with both conceptual and empirical outputs, the analysis of the temporal dynamics of metacommunities has only emerged recently and still consists mostly of r... |  | Biodiversity, Coexistence, Community ecology, Spatial ecology, Metacommunities & Metapopulations | Werner Ulrich | 2018-11-29 14:58:54 | View | |
12 May 2022

Riparian forest restoration as sources of biodiversity and ecosystem functions in anthropogenic landscapesYasmine Antonini, Marina Vale Beirao, Fernanda Vieira Costa, Cristiano Schetini Azevedo, Maria Wojakowski, Alessandra Kozovits, Maria Rita Silverio Pires, Hildeberto Caldas Sousa, Maria Cristina Teixeira Braga Messias, Maria Augusta Goncalves Fujaco, Mariangela Garcia Praca Leite, Joice Paiva Vidigal Martins, Graziella Franca Monteiro, Rodolfo Dirzo https://doi.org/10.1101/2021.09.08.459375Complex but positive diversity - ecosystem functioning relationships in Riparian tropical forestsRecommended by Werner Ulrich based on reviews by 2 anonymous reviewers based on reviews by 2 anonymous reviewers
Many ecological drivers can impact ecosystem functionality and multifunctionality, with the latter describing the joint impact of different functions on ecosystem performance and services. It is now generally accepted that taxonomically richer ecosystems are better able to sustain high aggregate functionality measures, like energy transfer, productivity or carbon storage (Buzhdygan 2020, Naeem et al. 2009), and different ecosystem services (Marselle et al. 2021) than those that are less rich. Antonini et al. (2022) analysed an impressive dataset on animal and plant richness of tropical riparian forests and abundances, together with data on key soil parameters. Their work highlights the importance of biodiversity on functioning, while accounting for a manifold of potentially covarying drivers. Although the key result might not come as a surprise, it is a useful contribution to the diversity - ecosystem functioning topic, because it is underpinned with data from tropical habitats. To date, most analyses have focused on temperate habitats, using data often obtained from controlled experiments. The paper also highlights that diversity–functioning relationships are complicated. Drivers of functionality vary from site to site and each measure of functioning, including parameters as demonstrated here, can be influenced by very different sets of predictors, often associated with taxonomic and trait diversity. Single correlative comparisons of certain aspects of diversity and functionality might therefore return very different results. Antonini et al. (2022) show that, in general, using 22 predictors of functional diversity, varying predictor subsets were positively associated with soil functioning. Correlational analyses alone cannot resolve the question of causal link. Future studies should therefore focus on inferring precise mechanisms behind the observed relationships, and the environmental constraints on predictor subset composition and strength. References Antonini Y, Beirão MV, Costa FV, Azevedo CS, Wojakowski MM, Kozovits AR, Pires MRS, Sousa HC de, Messias MCTB, Fujaco MA, Leite MGP, Vidigal JP, Monteiro GF, Dirzo R (2022) Riparian forest restoration as sources of biodiversity and ecosystem functions in anthropogenic landscapes. bioRxiv, 2021.09.08.459375, ver. 3 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2021.09.08.459375 Buzhdygan OY, Meyer ST, Weisser WW, Eisenhauer N, Ebeling A, Borrett SR, Buchmann N, Cortois R, De Deyn GB, de Kroon H, Gleixner G, Hertzog LR, Hines J, Lange M, Mommer L, Ravenek J, Scherber C, Scherer-Lorenzen M, Scheu S, Schmid B, Steinauer K, Strecker T, Tietjen B, Vogel A, Weigelt A, Petermann JS (2020) Biodiversity increases multitrophic energy use efficiency, flow and storage in grasslands. Nature Ecology & Evolution, 4, 393–405. https://doi.org/10.1038/s41559-020-1123-8 Marselle MR, Hartig T, Cox DTC, de Bell S, Knapp S, Lindley S, Triguero-Mas M, Böhning-Gaese K, Braubach M, Cook PA, de Vries S, Heintz-Buschart A, Hofmann M, Irvine KN, Kabisch N, Kolek F, Kraemer R, Markevych I, Martens D, Müller R, Nieuwenhuijsen M, Potts JM, Stadler J, Walton S, Warber SL, Bonn A (2021) Pathways linking biodiversity to human health: A conceptual framework. Environment International, 150, 106420. https://doi.org/10.1016/j.envint.2021.106420 Naeem S, Bunker DE, Hector A, Loreau M, Perrings C (Eds.) (2009) Biodiversity, Ecosystem Functioning, and Human Wellbeing: An Ecological and Economic Perspective. Oxford University Press, Oxford. https://doi.org/10.1093/acprof:oso/9780199547951.001.0001 | Riparian forest restoration as sources of biodiversity and ecosystem functions in anthropogenic landscapes | Yasmine Antonini, Marina Vale Beirao, Fernanda Vieira Costa, Cristiano Schetini Azevedo, Maria Wojakowski, Alessandra Kozovits, Maria Rita Silverio Pires, Hildeberto Caldas Sousa, Maria Cristina Teixeira Braga Messias, Maria Augusta Goncalves Fuja... | <ol> <li style="text-align: justify;">Restoration of tropical riparian forests is challenging, since these ecosystems are the most diverse, dynamic, and complex physical and biological terrestrial habitats. This study tested whether biodiversity ... |  | Biodiversity, Community ecology, Ecological successions, Ecosystem functioning, Terrestrial ecology | Werner Ulrich | 2021-09-10 10:51:23 | View | |
27 Jan 2023
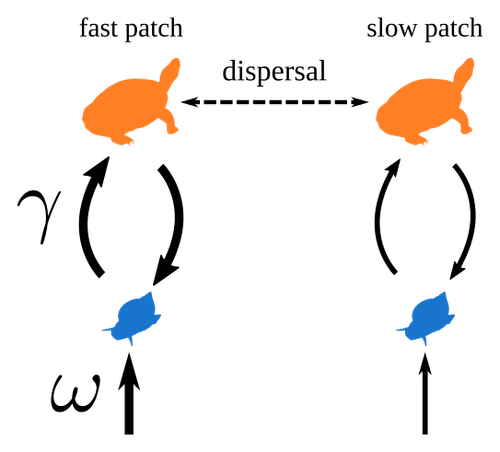
Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunitiesPierre Quévreux, Bart Haegeman and Michel Loreau https://hal.science/hal-03829838How does spatial heterogeneity affect stability of trophic metacommunities?Recommended by Werner Ulrich based on reviews by Phillip P.A. Staniczenko, Ludek Berec and Diogo Provete based on reviews by Phillip P.A. Staniczenko, Ludek Berec and Diogo Provete
The temporal or spatial variability in species population sizes and interaction strength of animal and plant communities has a strong impact on aggregate community properties (for instance biomass), community composition, and species richness (Kokkoris et al. 2002). Early work on spatial and temporal variability strongly indicated that asynchronous population and environmental fluctuations tend to stabilise community structures and diversity (e.g. Holt 1984, Tilman and Pacala 1993, McCann et al. 1998, Amarasekare and Nisbet 2001). Similarly, trophic networks might be stabilised by spatial heterogeneity (Hastings 1977) and an asymmetry of energy flows along food chains (Rooney et al. 2006). The interplay between temporal, spatial, and trophic heterogeneity within the meta-community concept has got much less interest. In the recent preprint in PCI Ecology, Quévreux et al. (2023) report that Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunities. These authors rightly notice that the interplay between trophic and spatial heterogeneity might induce contrasting effects depending on the internal dynamics of the system. Their contribution builds on prior work (Quévreux et al. 2021a, b) on perturbed trophic cascades. I found this paper particularly interesting because it is in the, now century-old, tradition to show that ecological things are not so easy. Since the 1930th, when Nicholson and Baily and others demonstrated that simple deterministic population models might generate stability and (pseudo-)chaos ecologists have realised that systems triggered by two or more independent processes might be intrinsically unpredictable and generate different outputs depending on the initial parameter settings. This resembles the three-body problem in physics. The present contribution of Quévreux et al. (2023) extends this knowledge to an example of a spatially explicit trophic model. Their main take-home message is that asymmetric energy flows in predator–prey relationships might have contrasting effects on the stability of metacommunities receiving localised perturbations. Stability is context dependent. Of course, the work is merely a theoretical exercise using a simplistic trophic model. It demands verification with field data. Nevertheless, we might expect even stronger unpredictability in more realistic multitrophic situations. Therefore, it should be seen as a proof of concept. Remember that increasing trophic connectance tends to destabilise food webs (May 1972). In this respect, I found the final outlook to bioconservation ambitious but substantiated. Biodiversity management needs a holistic approach focusing on all aspects of ecological functioning. I would add the need to see stability and biodiversity within an evolutionary perspective. References Amarasekare P, Nisbet RM (2001) Spatial Heterogeneity, Source‐Sink Dynamics, and the Local Coexistence of Competing Species. The American Naturalist, 158, 572–584. https://doi.org/10.1086/323586 Hastings A (1977) Spatial heterogeneity and the stability of predator-prey systems. Theoretical Population Biology, 12, 37–48. https://doi.org/10.1016/0040-5809(77)90034-X Holt RD (1984) Spatial Heterogeneity, Indirect Interactions, and the Coexistence of Prey Species. The American Naturalist, 124, 377–406. https://doi.org/10.1086/284280 Kokkoris GD, Jansen VAA, Loreau M, Troumbis AY (2002) Variability in interaction strength and implications for biodiversity. Journal of Animal Ecology, 71, 362–371. https://doi.org/10.1046/j.1365-2656.2002.00604.x May RM (1972) Will a Large Complex System be Stable? Nature, 238, 413–414. https://doi.org/10.1038/238413a0 McCann K, Hastings A, Huxel GR (1998) Weak trophic interactions and the balance of nature. Nature, 395, 794–798. https://doi.org/10.1038/27427 Quévreux P, Barbier M, Loreau M (2021) Synchrony and Perturbation Transmission in Trophic Metacommunities. The American Naturalist, 197, E188–E203. https://doi.org/10.1086/714131 Quévreux P, Pigeault R, Loreau M (2021) Predator avoidance and foraging for food shape synchrony and response to perturbations in trophic metacommunities. Journal of Theoretical Biology, 528, 110836. https://doi.org/10.1016/j.jtbi.2021.110836 Quévreux P, Haegeman B, Loreau M (2023) Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunities. hal-03829838, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://hal.science/hal-03829838 Rooney N, McCann K, Gellner G, Moore JC (2006) Structural asymmetry and the stability of diverse food webs. Nature, 442, 265–269. https://doi.org/10.1038/nature04887 Tilman D, Pacala S (1993) The maintenance of species richness in plant communities. In: Ricklefs, R.E., Schluter, D. (eds) Species Diversity in Ecological Communities: Historical and Geographical Perspectives. University of Chicago Press, pp. 13–25. | Spatial heterogeneity of interaction strength has contrasting effects on synchrony and stability in trophic metacommunities | Pierre Quévreux, Bart Haegeman and Michel Loreau | <p> Spatial heterogeneity is a fundamental feature of ecosystems, and ecologists have identified it as a factor promoting the stability of population dynamics. In particular, differences in interaction strengths and resource supply between pa... |  | Dispersal & Migration, Food webs, Interaction networks, Spatial ecology, Metacommunities & Metapopulations, Theoretical ecology | Werner Ulrich | 2022-10-26 13:38:34 | View | |
23 Oct 2023
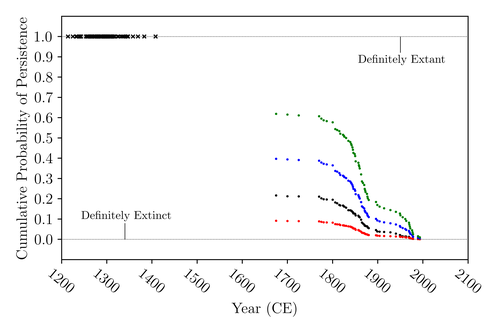
The Moa the Merrier: Resolving When the Dinornithiformes Went ExtinctFloe Foxon https://doi.org/10.1101/2023.08.07.552261Are Moas ancient Lazarus species?Recommended by Werner Ulrich based on reviews by Tim Coulson and Richard Holdaway based on reviews by Tim Coulson and Richard Holdaway
Ancient human colonisation often had catastrophic consequences for native fauna. The North American Megafauna went extinct shortly after humans entered the scene and Madagascar suffered twice, before 1500 CE and around 1700 CE after the Malayan and European colonisation. Maoris colonised New Zealand by about 1300 and a century later the giant Moa birds (Dinornithiformes) sharply declined. But did they went extinct or are they an ancient example of Lazarus species, species thought to be extinct but still alive? Scattered anecdotes of late sightings of living Moas even up to the 20th century seem to suggest the latter. The quest for later survival has also a criminal aspect. Who did it, the Maoris or the white colonisers in the late 18th century? The present work by Floe Foxon (2023) tries to settle this question. It uses a survival modelling approach and an assessment of the reliability of nearly 100 alleged sightings. The model favours the so-called overkill hypothesis, that Moas probably went extinct in the 15th century shortly after Maori colonisation. A small but still remarkable probability remained for survival up to 1770. Later sightings turned out to be highly unreliable. The paper is important as it does not rely on subjective discussions of late sightings but on a probabilistic modelling approach with sensitivity testing prior applied to marsupials. As common in probabilistic approaches, the study does not finally settle the case. A probability of as much as 20% remained for late survival after 1450 CE. This is not improbable as New Zealand was sufficiently unexplored in those days to harbour a few refuges for late survivors. However, in this respect, it is a bit unfortunate that at the end of the discussion, the paper cites Heuvelmans, the founder of cryptozoology, and it mentions the ivory-billed woodpecker, which has recently been redetected. No Moa remains were found after 1450. References Foxon F (2023) The Moa the Merrier: Resolving When the Dinornithiformes Went Extinct. bioRxiv, 2023.08.07.552261, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2023.08.07.552261 | The Moa the Merrier: Resolving When the Dinornithiformes Went Extinct | Floe Foxon | <p style="text-align: justify;">The Moa (Aves: Dinornithiformes) are an extinct group of the ratite clade from New Zealand. The overkill hypothesis asserts that the first New Zealand settlers hunted the Moa to extinction by 1450 CE, whereas the st... |  | Conservation biology, Human impact, Statistical ecology, Zoology | Werner Ulrich | Tim Coulson, Richard Holdaway | 2023-08-08 17:14:30 | View |
MANAGING BOARD
Julia Astegiano
Tim Coulson
Anna Eklof
Dominique Gravel
François Massol
Ben Phillips
Cyrille Violle










