
Ensuring reproducible science requires policies
Code-sharing policies are associated with increased reproducibility potential of ecological findings
Recommendation: posted 26 March 2025, validated 26 March 2025
Bartomeus, I. (2025) Ensuring reproducible science requires policies. Peer Community in Ecology, 100778. https://doi.org/10.24072/pci.ecology.100778
Recommendation
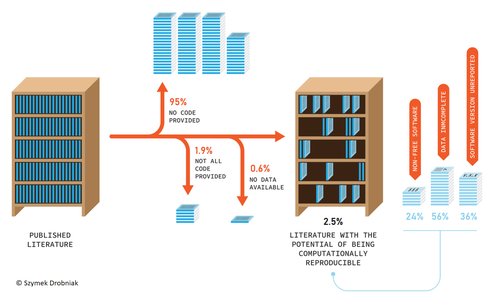
Researchers do not live in a vacuum, and the social context we live in affects how we do science. On one hand, increased competition for scarce funding creates the wrong incentives to do fast analysis, leading sometimes to poorly checked results that accumulate errors (Fraser et al. 2018). On the other hand, the actual challenges the world faces require more than ever robust scientific evidence that can be used to tackle the current rapid human-induced environmental change. Moreover, scientists' credibility is at stake at this moment where the global flow of information can be politically manipulated, and accessing reliable sources of information is paramount for society. At the crossroads of these challenges is scientific reproducibility. Making our results transparent and reproducible ensures that no perverse incentives can compromise our findings, that results can be reliably applied to solve relevant problems, and that we regain societal credibility in the scientific process. Unfortunately, in ecology and evolution, we are still far from publishing open, transparent, and reproducible papers (Maitner et al. 2024). Understanding which factors promote increased use of good practices regarding reproducibility is hence very welcome.
Sanchez-Tojar and colleagues (2025) conducted a (reproducible) analysis of code and data-sharing practices (a cornerstone of scientific reproducibility) in journals with and without explicit policies regarding data and code deposition. The gist is that having policies in place increases data and code sharing. Doing science about how we do science (meta-science) is important to understand which actions drive our behavior as scientists. This paper highlights that in the absence of strong societal or personal incentives to share code and data, clear policies can catalyze this process. However, in my opinion, policies are a needed first step to consolidate a more permanent change in researchers' behavior regarding reproducible science, but policies alone will not be enough to fix the problem if we do not change also the cultural values around how we publish science. Appealing to inner values, and recognizing science needs to be reproducible to ensure potential errors are easily spotted and corrected requires a deep cultural change.
References
Fraser, Hannah, Tim Parker, Shinichi Nakagawa, Ashley Barnett, and Fiona Fidler. "Questionable research practices in ecology and evolution." PloS one 13, no. 7 (2018): e0200303. https://doi.org/10.1371/journal.pone.0200303
Maitner, Brian, Paul Efren Santos Andrade, Luna Lei, Jamie Kass, Hannah L. Owens, George CG Barbosa, Brad Boyle et al. "Code sharing in ecology and evolution increases citation rates but remains uncommon." Ecology and Evolution 14, no. 8 (2024): e70030. https://doi.org/10.1002/ece3.70030
Alfredo Sánchez-Tójar, Aya Bezine, Marija Purgar, Antica Culina (2025) Code-sharing policies are associated with increased reproducibility potential of ecological findings. EcoEvoRxiv, ver.4 peer-reviewed and recommended by PCI Ecology. https://doi.org/10.32942/X21S7H
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Corona-Unterstützungsupport program, Bielefeld University; Croatian Science Foundation (HRZZ-IP-2022-10-2872; DOK-2021-02-6688); Fulbright U.S. Student Program.
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.32942/X21S7H
Version of the preprint: 1
Author's Reply, 26 Mar 2025
Decision by Ignasi Bartomeus , posted 06 Feb 2025, validated 07 Feb 2025
, posted 06 Feb 2025, validated 07 Feb 2025
I welcome this formal analysis of reproducibility practices in Ecology. In general, I don't have any major concerns with the paper, but both reviewers raise important points that need to be addressed before recommending the manuscript. Some of those comments refer to issues that might not be easily fixed, such as the relatively smaller sample size or the fact that the study does not cover more recent time periods. Still, a good discussion should be included regarding those points. However, other points, such as the revision of the supplementary material of the analyzed manuscripts, are appropriate and will reinforce the message of the paper.
I am looking forward to reading an enhanced version of the paper.
Best,
Ignasi Bartomeus.
Reviewed by Francisco Rodriguez-Sanchez , 21 Jan 2025
, 21 Jan 2025
Title and abstract
Does the title clearly reflect the content of the article? YES
Does the abstract present the main findings of the study? YES
Introduction
Are the research questions/hypotheses/predictions clearly presented? YES
Does the introduction build on relevant research in the field? YES
Materials and methods
Are the methods and analyses sufficiently detailed to allow replication by other researchers? YES (although the list of examined papers is not provided)
Are the methods and statistical analyses appropriate and well described? YES, but see comments below
Results
Are the results described and interpreted correctly? YES, but see comments below
Discussion
Have the authors appropriately emphasized the strengths and limitations of their study/theory/methods/argument? YES
Are the conclusions adequately supported by the results (without overstating the implications of the findings)? YES
------------------------------------------------------------------------------------------------------------------
Sánchez-Tójar et al. present an interesting analysis of how journal policies on code sharing may affect levels and quality of actual code sharing, determining the degree of computational reproducibility of published scientific results in ecology. This is an important topic and a nice follow-up from Culina et al. (2020) which analysed the computational reproducibility in ecological journals encouraging or mandating code sharing. Here, those results are contrasted against a comparable sample of ecological journals without a code sharing policy, finding that, in fact, journals' code sharing policies seem to increase actual code sharing and computational reproducibility.
The comparison with Culina et al. (2020) implies that papers from the same years (2015 to 2019) were analysed here. More than five years have passed now, which is a long time in the open science world, in the sense that actual code sharing practices may have changed now, and be significantly different from the figures reported here (from a few years ago). This is fine and justified by the planned comparison with Culina et al. (2020), with the main goal of assessing the impact of journal policies on code sharing. Other pieces of work (e.g. Maitner et al. 2024 https://doi.org/10.1002/ece3.70030, and others to come) may provide more updated assessments of code sharing rates and quality.
Main comments
(1) L161-162: "we searched for software and package versions not only within the text but also in the reference list of the corresponding article".
I think this methodology may be underestimating the numbers on the reporting of software and package versions, as I think many articles report that information in the Supplementary Material (e.g. Extended Methods section, or Software Appendix), or even within the archived code, rather than within the main text or reference list. Hence, I think it may be important to revise at least the Supplementary Information of all the examined papers (where existing) to check if software and packages are reported there. To be fully comprehensive, provided code should be revised as well, for the few papers that provide it. Numbers might not change much compared to current figures, but I am afraid we will not know unless we check.
(2) L290-303: "We found that versions of the statistical software and packages were often missing, and about a tenth of the articles did not even state the software used. Reporting software and package versions is important for several reasons. First, they can help in understanding and solving technical issues related to software dependencies, which are one of the most often encountered factors hindering computational reproducibility (Laurinavichyute et al. 2022; Kellner et al. 2024; Samuel and Mietchen 2024). Different versions of software and/or packages can lead to inconsistencies in results and even to code rot, which occurs when code relies on specific versions of software or packages that are no longer available or have undergone significant changes (e.g., deprecated functions), rendering the code incompatible with current operating systems (Boettiger 2015; Laurinavichyute et al. 2022)."
This is related to the comment above. The authors seem to expect (and recommend) that software and package versions are reported in the main text and reference list to guarantee computational reproducibility. But is it feasible or reasonable to expect that all software dependencies are cited in the main text, to guarantee computational reproducibility? A typical analysis in ecology may involve dozens, or even more than a hundred, software dependencies (including indirect dependencies).
Current guidelines for software citation (Chue Hong et al. 2019, https://doi.org/10.5281/zenodo.3479198) state that "you should cite software that has a significant impact on the research outcome presented in your work, or on the way the research has been conducted. In general, you do not need to cite software packages or libraries that are not fundamental to your work and that are a normal part of the computational and scientific environment used. These dependencies do not need to be cited outright but should be documented as part of the computational workflow for complete reproducibility".
Thus, the main function of software citations in the reference list is not to guarantee computational reproducibility, but to explain methods and give credit to software developers. For the sake of computational reproducibility, a better way of reporting dependencies is including a full report of the computational environment used for the analyses, including, of course, package versions. It would be nice, in my opinion, if this manuscript included brief guidelines on how to do so within the final recommendations, or at least point to useful tutorials. For example, focusing on R, authors could include an appendix generated with `sessionInfo()` or `grateful::cite_packages()`, or include in their code a file reporting the computational environment (e.g. a Dockerfile, or at least a `renv.lock` file generated with `renv::snapshot()`). In any case, I think the Discussion should better clarify the different roles of software citation (credit, reproducibility, etc) and revise their recommendations accordingly.
(3) Fig. 2 seems confusing. In panels a and b, the 'No' category appears in black in the bottom. But in other panels it is the 'Yes' category which appears in black in the bottom. I would suggest to revise the figure design to make it more consistent across panels.
Other comments
(4) L73-76: "Although container technology such as Docker, which packages the software and its dependencies into a standardized environment, has been suggested as a solution to improving portability and reproducibility, its adoption remains low".
I share the same impression about low adoption of Docker among researchers, but is there any study or numbers to support that statement? For example, how many articles sharing code include a Dockerfile? I do not know of specific studies, but I think it would be useful to cite them if available. Maybe Carl Boettiger may know? (cf. https://doi.org/10.1038/s43586-023-00236-9)
(5) L83-86: "code should be shared in a permanent repository (e.g., Zenodo) and assigned with an open and permissive licence and a persistent identifier such as a DOI. This is particularly important given the far-from-ideal rates of link persistence found for scientific code in fields such as astrophysics (Allen et al. 2018)".
Sperandii et al. 2024 (https://doi.org/10.1111/jvs.13224) state that "For code, the most frequent reason for inaccessible code was a broken link (69.2%), whereas the use of private repositories was much less common (7.7%)". Perhaps they could be cited here to support that statement.
(6) L150 and Fig. 2: better explain what 'partially free' software means? Is that a combination of free and propietary software within the same paper?
(7) L193-198: This is fine as is, but I wonder if, for the sake of clarity, these results would rather be reported in a positive framing, e.g. rather than "36.1% (N = 100) of articles published in journals without a code sharing policy did not report the version of all software used", write "63.9% (N = XXX) of articles published in journals without a code sharing policy reported the version of all software used". Sounds less convoluted. Applies also to L210-212.
(8) L213-214: "The mean number of packages used was 2.30 (median = 2.00, range = 1 to 10) in journals without a code sharing policy and 2.41 (median = 2.00, range = 1 to 14) in journals with a code-sharing policy". As far as I understand, this is the number of packages cited in the main text. The actual number of packages used in the analyses is probably quite larger. Please rephrase.
(9) L245-246: "several transparency indicators, including, but not limited to, data- and code-sharing are on the rise in ecology (Evans 2016; Culina et al. 2020; Roche et al. 2022a)". Include results from Maitner et al 2024 here too (https://doi.org/10.1002/ece3.70030)?
References
Boettiger et al. 2015 reference is missing
Update Ivimey-Cook et al (in prep) to preprint reference?
https://doi.org/10.24072/pci.ecology.100778.rev11Reviewed by Veronica Cruz, 06 Feb 2025
In my review I have evaluated the main points recommended by PCIEcol. In addition, I attach the manuscript in pdf with some minor comments in case the authors want to consider them. I hope the comments are useful and I apologize for my late answer.
Title
Check that the title clearly reflects the content of the article.
It does.
Abstract
Check that the abstract is concise and presents the main findings of the study.
It does.
Introduction
Check that the introduction clearly explains the motivation for the study. Check that the research question/hypothesis/prediction is clearly presented. Check that the introduction builds on relevant recent and past research performed in the field.
To my understading, the introduction explains the motivation of the study and presents clear objectives and hypotheses. However, the results and discussion go beyond these objectives. The novelty of the study is not so clear. The authors cite other articles (even authored by them*) that cover very similar topics. I am not very familiar with the literature in the field but it seems updated and rich.
*The article Ivimey-Cook et al. in prep is cited in the introduction and discussion several times. I find it inappropriate since an article in preparation may end up with very different conclusions. I encourage the authors to upload a preprint of this article if they want to use it to support the manuscript or use it only to support very specific statements of the manuscript.
Materials and methods
More generally, check that sufficient details are provided for the methods and analysis to allow replication by other researchers.
I was surprised that given the topic of the article, the authors did not mention in the main text that data management and analysis were done using R and which were the main packages used. I think this is mandatory even though they provide scripts with all the necessary information.
There are several decisions on methodology that seem arbitrary to me, and I think they need further explanation. Although authors followed a methodology that makes results comparable to those in Culina et al. 2020 and thus they refer frequently to this article, some important decisions should be briefly justified in the current article as well. For example, I don’t think that separating data in two periods is fulfilling the objectives stated by authors here.
Also, I appreciate the honesty of saying that studies with landscape analysis were discarded because authors “lacked the expertise to understand the analyses and software used.”. However, what is the impact of discarding landscape (and molecular analysis as well)? They might be discarding a large number of articles from journals such as Landscape Ecology. If the authors assume that these journals are representative of ecology, why discard certain disciplines? If the scope of the review is missing some parts, it should be clearly stated in results/conclusions/summary.
Check that the statistical analyses are appropriate.
They are exploratory analysis, and I find them appropriate.
Results
I think the results did not exactly address the objectives. They extensively described the results for journals without code-sharing policies, but they only make a comparison at the end of the first results section (L189) with journals with code-sharing policies. I think that results should be more focused in comparison and maybe report results for journals without code-sharing policies in supplementary materials. They did not state any objective or hypothesis related to different time periods, yet they extensively reported differences between two periods in text and figures and discussed it deeply.
If possible, evaluate the consistency of raw data and scripts.
All literature reviews or meta-analysis that I know analyze at least the title and abstract of the entire set of articles given by the literature search. I don’t think that analyzing a sample of less than 10% of the entire population of the bibliography can make results generalizable. I recommend either expanding the sample of analyzed articles (in journals with and without code-sharing policies) or acknowledging this limitation and including the impact of this decision in the discussion of the results and in conclusions. The subset of articles with available code is very small and could substantially change if another random sample of articles is studied.
In script 003_plotting, the ggplot2 and patchwork libraries need to be explicitly loaded. I have some problems with long directory addresses and file names.
If necessary, and if you can, run the data transformations and statistical analyses and check that you get the same results. In the case of negative results, check that there is a statistical power analysis (or an adequate Bayesian analysis).
I did not check all the results, but the ones I did were consistent.
Inform the recommender and the managing board if you suspect scientific misconduct.
I did not suspect scientific misconduct.
Tables and figures
Check that figures and tables are understandable without reference to the main body of the article. Check that figures and tables have a proper caption.
In Table 1, I don’t understand what is “[using data]”. Also, here and in other parts of the manuscript, there is no need to say “nonmolecular” articles.
Figure 1 is clear.
Figure 2 can be improved. The dotted pattern is distractive and not needed. Also, it is confusing that sometimes “yes” is white and sometimes is black. I recommend, if possible, to use same color for the desirable output (i.e. reported software, reported versions, free software) all the time. It is also confusing that the caption does not first explain the first panel.
Discussion
Check that the conclusions are adequately supported by the results and that the interpretation of the analysis is not overstated.
I think conclusions are adequate, except that the results do not support calling for funders to introduce code- and data-sharing policies.
Check that the discussion takes account of relevant recent and past research performed in the field.
I am not very familiar with literature in the field. As a general comment I think that the discussion is biased towards temporal trends in code-sharing rather than the differences between journals with and without code-sharing policies.
Download the review https://doi.org/10.24072/pci.ecology.100778.rev12









