
Transcriptomics of thermal stress response in corals
Gene expression plasticity and frontloading promote thermotolerance in Pocillopora corals
Abstract
Recommendation: posted 26 August 2019, validated 04 September 2019
Jacob, S. (2019) Transcriptomics of thermal stress response in corals. Peer Community in Ecology, 100028. 10.24072/pci.ecology.100028
Recommendation
Climate change presents a challenge to many life forms and the resulting loss of biodiversity will critically depend on the ability of organisms to timely respond to a changing environment. Shifts in ecological parameters have repeatedly been attributed to global warming, with the effectiveness of these responses varying among species [1, 2]. Organisms do not only have to face a global increase in mean temperatures, but a complex interplay with another crucial but largely understudied aspect of climate change: thermal fluctuations. Understanding the mechanisms underlying adaptation to thermal fluctuations is thus a timely and critical challenge.
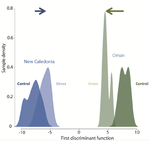
Coral reefs are among the most threaten ecosystems in the context of current global changes [3]. Brener-Raffalli and colleagues [4] provided a very complete study digging into the physiological, symbiont-based and transcriptomic mechanisms underlying response of corals to temperature changes. They used an experimental approach, following the heat stress response of coral colonies from different species of the genus Pocillopora. While the symbiont community composition did not significantly change facing exposure to warmer temperatures, the authors provided evidence for transcriptomic changes especially linked to stress response genes that may underlie plastic responses to heat stress.
The authors furthermore investigated the thermal stress response of corals originating from two sites differing in their natural thermal regimes, and found that they differ in the extent and nature of plastic response, including the expression of gene regulation factors and the basal expression level of some genes. These two sites also differ in a variety of aspects, including the focal coral species, which precludes from concluding about the role of thermal regime adaptation into the differences observed. However, these results still highlight a very interesting and important direction deserving further investigation [5], and point out the importance of variability in thermal stress response among localities [6] that might potentially mediate global warming consequences on coral reefs.
References
[1] Parmesan, C., & Yohe, G. (2003). A globally coherent fingerprint of climate change impacts across natural systems. Nature, 421(6918), 37–42. doi: 10.1038/nature01286
[2] Menzel, A., Sparks, T. H., Estrella, N., Koch, E., Aasa, A., Ahas, R., … Zust, A. (2006). European phenological response to climate change matches the warming pattern. Global Change Biology, 12(10), 1969–1976. doi: 10.1111/j.1365-2486.2006.01193.x
[3] Bellwood, D. R., Hughes, T. P., Folke, C., & Nyström, M. (2004). Confronting the coral reef crisis. Nature, 429(6994), 827–833. doi: 10.1038/nature02691
[4] Brener-Raffalli, K., Vidal-Dupiol, J., Adjeroud, M., Rey, O., Romans, P., Bonhomme, F., Pratlong, M., Haguenauer, A., Pillot, R., Feuillassier, L., Claereboudt, M., Magalon, H., Gélin, P., Pontarotti, P., Aurelle, D., Mitta, G. and Toulza, E. (2019). Gene expression plasticity and frontloading promote thermotolerance in Pocillopora corals. BioRxiv, 398602, ver 4 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/398602
[5] Kenkel, Carly D., and Matz, M. V. (2017). Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nature Ecology and Evolution, 1(1), 0014. doi: 10.1038/s41559-016-0014
[6] Kenkel, C. D., Meyer, E., and Matz, M. V. (2013). Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Molecular Ecology, 22(16), 4322–4334. doi: 10.1111/mec.12390
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/398602
Version of the preprint: 2
Author's Reply, 20 Aug 2019
Decision by Staffan Jacob, posted 17 May 2019
Dear Eve Toulza,
following the submissions of your revisions, the two reviewers and myself agree to acknowledge helpful clarifications of several aspects of your manuscript. The reviewers still pointed out a number issues that should be dealt with, especially regarding the introduced concepts on the link between plasticity and adaptation and the way analyses were performed. I thus invite you to carefully use the reviewers’ comments to revise your manuscript.
All the best, Staffan.
Additional requirements of the managing board:
As indicated in the 'How does it work?’ section and in the code of conduct, please make sure that:
-Data are available to readers, either in the text or through an open data repository such as Zenodo (free), Dryad (to pay) or some other institutional repository. Data must be reusable, thus metadata or accompanying text must carefully describe the data.
-Details on quantitative analyses (e.g., data treatment and statistical scripts in R, bioinformatic pipeline scripts, etc.) and details concerning simulations (scripts, codes) are available to readers in the text, as appendices, or through an open data repository, such as Zenodo, Dryad or some other institutional repository. The scripts or codes must be carefully described so that they can be reused.
-Details on experimental procedures are available to readers in the text or as appendices.
-Authors have no financial conflict of interest relating to the article. The article must contain a "Conflict of interest disclosure" paragraph before the reference section containing this sentence: "The authors of this preprint declare that they have no financial conflict of interest with the content of this article." If appropriate, this disclosure may be completed by a sentence indicating that some of the authors are PCI recommenders: “XXX is one of the PCI XXX recommenders.”
Reviewed by Mar Sobral, 16 Apr 2019
I already pointed out in my previous revision that this work is very interesting and important. The writing is also very good. But, although improvement has been made to the manuscript in comparison to previous version, I still disagree with a couple of important conceptual as well as analytical issues (that I had already pointed out in my previous revision). So, my recommendation is again a major revision. This I because I believe there is conceptual confusion that wont help understanding the implications of the work. Second, the analytical limitations preclude the exploitation of results to their maximum potential.
CONCEPTUAL ISSUES
Line 43-44. It is not possible to interpret this work in the context of differences between localities, and I , the other referee and the editor explicitly said in previous version. So, line 40-41 is not what is tested in this research. Same for lines 79-80.
Line 83-94. In my opinion this is wrong, as I already explained in my previous revision. Refocuse it more in line of what you say in lines 94-97.
Line 98-100. No, you cannot discriminate between both processes with molecular approaches. And in any case, both would be entangled. Also, you have not data to test whether local adaptation is going on etc.
Line 114-117. That is right, and that explains my two previous comments.
Line 191-196. This is a good prediction to make, but a different story is to say that testing that is the purpose of the research (because in that case design would be not ok).
ANALYTICAL ISSUES
Line 334, performing one-way anovas for alpha diversity is not correct here, as I said in my previous revision. You have 42 alpha values right? Therefore, including site, colony within site, treatment and treatment*site should be possible and they need to be factors explaining alpha diversity at the colony level. Maybe you can assess how stress affects beta diversity whether you asses beta diversity within colonies (among replicates) and then you can analyze it as a response variable including treatment, site and their interaction in the model (if I don’t misunderstand u can have n=12 values of beta diversity, because you can asses it among replicates within colonies and different betas for different teratments.). If I am wrong, please explain why and make it clear in the manuscript that no such an analyses are possible to run.
MINOR COMMENTS
Line 160-161. You do not test the role of microorganisms for this, is misleading mentioning it here?
L 238 and 244. You say 8 tanks per colony first, and later 4. I understand is 4.
Line 388 and 392, change condition for “treatment” .
Line 414. Then, what is the name of this species? Again, this kind of means that design mixes species with localities, that is yet another reason NOT to interpret results in function of localities differences, but rather only in function of treatment differences.
Line 417. Given this result is would make more sense to analyze either NC2 or NC3 all throughout the paper. It doesn’t make a lot of sense to have a replicated genotype but no others. It is likely that differences between NC2 and NC3 can support some interpretations, in comparison with differences among other colonies (I am not thinking about anything in particular right now).
Lines 443-445. This needs to be tested trough stats. I had already pointed this out in my previous revision.
Line 449-455. This analysis is not correct, as I pointed out in a previous comment.
Line 462-468. Why haven’t you analyzed this at the OUT level? And just used “types”?
Figure 7. I believe it is best to display it by colony. In the same colors and shades but making the differences between colonies.
Line 536-538. Cool!
Line 736-737. I remain unsure about these results as in previous versions.
Line 749-751. Empty sentences as it is. Remove or develop if important.
Line 7454-757. This is not necessary to be evolutionary meaningful. As I had already pointed out in my previous revision with references included (non-genetic inheritance affecting evolutionary dinamics).
Line 819 Change line for row.
Fig 6. Isn’t it misleading? Shouldn’t be all genes plotted and later, differences in density between quadrants could tell us whether there are more genes which behave similarly or differently between localities etc?
*I think it is necessary to explain how DEseQ2 works
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/398602
Version of the preprint: 1
Author's Reply, 08 Apr 2019
Decision by Staffan Jacob, posted 26 Oct 2018
Dear authors,
two reviewers have read your manuscript entitled "Gene expression plasticity and frontloading promote thermotolerance in Pocilloporid corals" and provided detailed and constructive reviews. As you will see, they both appreciated the topic, the experimental approach used and the impressive diversity of measures performed. However, they also raised a number of important concerns and requests, from the theory concepts to clarification of experimental design and important changes into the way analyses were performed. The authors should also modify throughout the manuscript the statements made about the link between thermal sensitivity and the traits quantified to make clear that the study consist in contrasting two populations (and even species), that differ in thermal regime but certainly not only. I need to make clear that, although your manuscript potentially represent an interesting contribution to the literature, that the conclusions hold on a comparison between two populations/species is a critical specificity of this study that should be outlined and made clear throughout the manuscript.
I encourage the authors to consider these detailed reviews to carefully revise their manuscript. Please keep in mind that the correct integration of the reviewers' requests is compulsory for the acceptance of this manuscript, but that I am uncertain at this step whether all these serious objections can be adequately addressed.
Thank you for submitting your work to PCI Ecology. I look forward to see the revised version of your manuscript soon.
Sincerely Staffan
Reviewed by Mar Sobral, 19 Oct 2018
Comments by Mar Sobral on the manuscript submitted to PCI "Gene expression plasticity and frontloading promote thermotolerance in Pocilloporid corals" by Brener-Raffalli et al. and submitted by Eve Toulza
I believe that studying gene expression plasticity and frontloading in face of a realistic and important environmental change (heat stress), for corals (which sustain a big amount of marine diversity) is a wonderful task to undertake. Both, because of the fundamental implications of this research (advances on the knowledge of evolutionary processes) and because of the potential practical use of this information. Even more, the fact that these organisms are holobionts and that bacterial and algae communities associated to the cnidarian hosts were investigated is a wonderful characteristic of this study. I also commend the authors for using so many alternative molecular approaches for ascribing the hosts to the species level, studying gene expression via transcriptomes, and studying bacterial and algae communities’ compositions. Not only that, they used bioinformatics to search the proteins and function for each gene with a differential expression in the experiment. Additionally, I agree with the sampling and experimental design of the study. Because of all the previous reasons I believe this work has a huge potential. However, there are other reasons -which I believe are very important- to make me think that, before publication, this work should go trough a major revision. Particularly regarding data analyses and result interpretation. In summary; I believe that the work is really good but the manuscript is very immature yet. Here I explain some of my concerns.
Line 39 (and affecting the whole manuscript): With only 2 populations, each belonging to one of contrasting environments, this hypothesis is not tested. To test whether differences among populations from contrasting environments are due to environmental characteristics at the population level, it would be necessary to have several populations per contrasting environment or several populations along an environmental gradient (so to test a statistical relationship between environment and gene expression at the population level). Additionally, 3 different coral species were studied (one in Oman, with 3 colonies), another in New Caledonia (with one colony) and another in NC (with two colonies). These means that colony is not the same level of organization in OM and in NC, and that the comparison between NC and OM is not ok, because different coral species are being compared.
Lines 76-78 (and affecting the whole paper): The dilemma is not between acclimatization via intra generational plasticity and intergenerational Darwinian adaptation. As you say later, both processes can occur at the same time and complement each-other. But even more important is that there is also trans-generational plasticity, which is the clearer link between acclimatization and Darwinian microevolution. This is because natural selection acts on phenotypes, therefore transgenerational plasticity will change phenotypes available for natural selection to act upon, changing the evolutionary course of a population (see for example Day, Troy, and Russell Bonduriansky. "A unified approach to the evolutionary consequences of genetic and nongenetic inheritance." The American Naturalist 178.2 (2011): E18-E36, and other works by these authors). Additionally, the frontloading discussion (all over the manuscript) should have into account the “priming” processes explained in epigenetics (see works by Eva Jablonka). Regarding analyses of gene expression (L 393-400), I believe that expression differences between before and after the stress, should be the response variable in a model with locality, colony (nested within locality) and treatment and interaction between treatment and locality as factors. Another possibility is substitute localities and colonies by species, regrouping data in a different manner but probably the balance won’t be ok to do that.
I am not convinced that there were not differences between bacterial communities from hosts from different treatments. First, I believe that Fig. 2 needs to be changed in such a way that it is easier to understand. The legend and combination of colors and shapes makes it difficult to read. In any case, I can clearly see that for the first four genotypes in the legend there’s always one of the stressed samples with a very different bacterial community than others in same genotype. I believe that an additional analysis should be made to test that. I believe that it is also needed a statistical procedure linking distances among bacterial communities with gene expression differences in hosts from control and stressed samples. That result (if there is a relationship) would prove that plastic and epigenetic effects not only affect evolution of involved populations but shape entire communities (in an analogous manner of works about community genetics by Thomas G. Whittam but with epigenetics) which is a very important potential results that goes overlooked in the current version of the manuscript. Additionally, (L 260-262) I believe this analysis is wrong. Alfa and Beta diversity should be response variables in a model including simultaneously locality, colony nested within locality, treatment (and if power enough the interaction between locality and treatment). I don’t understand why one-way ANOVAS were chosen.
Some doubts about design: All the nubbins bleached at the same time? It would have been necessary to use survival rates as a fitness measure at a point during the experiment to measure the costs of frontloading and plasticity to fitness (as you say in L 578). Why didn´t you do that? Do you have the data to test that idea? In L 193-194 I see that four nubbins per colony were placed in each tank, right? That would mean there were 12 nubbins per tank (8 tanks total). So, there were 96 nubbins, why later only 36 were studied? (L. 208). Explanation about this is needed. From the nubbins remaining in the experiment, were the 3 stressed ones per colony and population belonging to different tanks? same tank? etc? I believe it is necessary to include a figure explaining the experimental design. (It is possible that some of my comments reflect a lack of perfect understanding of what exactly was done with samples and data. That would mean that the writing should be much clearer).
MINOR COMMENTS -I believe that a better explanation on how the Go analysis works is necessary, and also, that too much detail is given on that section, especially given that those results are partly speculative. I would move the function result section to supplementary materials (leaving only a general paragraph about patterns found) and the same is true for the functional section in the discussion. It makes the discussion unnecessarily long and takes space for potentially more interesting discussion on the evolution of plasticity (see works by Kathleen Donohue). For example, I would discuss a bit differences between predictable, variable and predictably variable environments and which strategies could be expected in each case. -Line 52: What do you mean by highly enriched? -Line 53: Advance which insights. -Line 99-100, I believe you mean via transgenerational epigenetics processes, right? -Line 86-87, typo? Something lacking? Sentence is not understandable. -Often, specially between text and figures you mix the term colony and genotype, please use the same throughout the manuscript. Same occurs for population and locality (Locality is changed for population in the supplementary materials, and supplementary materials are refereed to as additional materials). -L. 166: With this aim. -L. 181: How many clones? -L 186. As I said, no implications can be made from environmental differences because only two localities are studied, but even more important because different species are studied per locality and in NC ,2 different species are mixed. -L 45 and 203 seem contradictory. -L 295. I believe the use of a posteriori is wrong in this context -L 264. The comparison should be made per colony so that we avoid the problems of mixing different species and localities. -L 296. As I said, comparing localities with different species is not ok. -L414-415. From these lines I don´t see what the analysis was about. It seems wrong, unless I miss something. L 415-417. Having the result of the GLM why you would need to perform Wilcoxon tests. Again, I am not sure if I miss something. L 479. Which analysis? L504. I disagree, the experimental design doesn’t allow to focus results this way as I comment previously. Same for line 525. L 520. Remove to. L 556. This comparison should me made within genotype, not possible to mix the responses of different genotypes and specially such in the case of NC if they belong to different species. L568: Which stats were used for this comparison? L 586, Very few what? L 600. Eliminate also. L 628. I don’t understand what you mean, explain where it comes from please. L741-742. That is great, I would focus this entire section on this idea making it less descriptive and shorter. -Fig 6. In the text it is said that there are 24 genes which were overexpressed in OM and under-expressed in NC (L 406-407), why they aren´t show in Fig 6? Am I missing anything? -Fig 7. Interestingly the transcriptomes from stressed colonies were more like another than non-stressed transcriptomes, I believe that result is very relevant and I didn´t find anything about it in the text.










