
GRIFFITHS Megan
- School of Biodiversity One Health and Veterinary Medicine, University of Glasgow, Glasgow, United Kingdom
- Host-parasite interactions, Microbial ecology & microbiology, Molecular ecology, Phylogeny & Phylogeography, Population ecology, Statistical ecology
Recommendations: 0
Review: 1
Review: 1
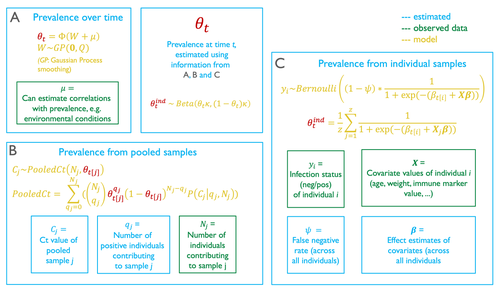
Reconstructing prevalence dynamics of wildlife pathogens from pooled and individual samples
Pooled samples hold information about the prevalence of wildlife pathogens
Recommended by Timothée Poisot based on reviews by Megan Griffiths and 2 anonymous reviewersAlthough monitoring the prevalence of pathogens in wildlife is crucial, there are logistical constraints that make this difficult, costly, and unpractical. This problem is often compounded when attempting to measure the temporal dynamics of prevalence. To improve the detection rate, a commonly used technique is pooling samples, where multiple individuals are analyzed at once. Yet, this introduces further potential biases: low-prevalence samples are effectively diluted through pooling, creating a false negative risk; negative samples are masked by the inclusion of positive samples, possibly artificially inflating the estimate of prevalence (and masking the inter-sample variability).
In their contribution, Borremans et al. (2024) come up with a modelling technique to provide accurate predictions of prevalence dynamics using a mix of pooled and individual samples. Because this model represents the pooling of individual samples as a complete mixing process, it can accurately estimate the prevalence dynamics from pooled samples only.
It is particularly noteworthy that the model provides an estimation of the false negative rate of the test. When there are false negatives (or more accurately, when the true rate at which false negatives happens), the value of the effect coefficients for individual-level covariates are likely to be off, potentially by a substantial amount. But besides more accurate coefficient estimation, the actual false negative rate is important information about the overall performance of the infection test.
The model described in this article also allows for a numerical calculation of the probability density function of infection. It is worth spending some time on how this is achieved, as I found the approach relying on combinatorics to be particularly interesting. When pooling, both the number of individuals that are mixed is known, and so is the measurement made on the pooled samples. The question is to figure out the number of individuals that because they are infectious, contribute to this score. The approach used by the authors is to draw (with replacement) possible positive and negative test outcomes assuming a number of positive individuals, and from this to estimate a pathogen concentration in the positive samples. This pathogen concentration can be transformed into its test outcome, and this value taken over all possible combinations is a conditional estimate of the test outcome, knowing the number of pooled individuals, and estimating the number of positive ones.
This approach is where the use of individual samples informs the model: by providing additional corrections for the relative volume of sample each individual provides, and by informing the transformation of test values into virus concentrations.
The authors make a strong case that their model can provide robust estimates of prevalence even in the presence of common field epidemiology pitfalls, and notably incomplete individual-level information. More importantly, because the model can work from pooled samples only, it gives additional value to samples that would otherwise have been discarded because they did not allow for prevalence estimates.
References
Benny Borremans, Caylee A. Falvo, Daniel E. Crowley, Andrew Hoegh, James O. Lloyd-Smith, Alison J. Peel, Olivier Restif, Manuel Ruiz-Aravena, Raina K. Plowright (2024) Reconstructing prevalence dynamics of wildlife pathogens from pooled and individual samples. bioRxiv, ver.3 peer-reviewed and recommended by PCI Ecology https://doi.org/10.1101/2023.11.02.565200