
Urban past predicts contemporary genetic structure in city rats
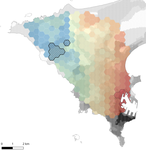
Interplay between historical and current features of the cityscape in shaping the genetic structure of the house mouse (Mus musculus domesticus) in Dakar (Senegal, West Africa)
Abstract
Recommendation: posted 11 May 2020, validated 11 May 2020
DiLeo, M. (2020) Urban past predicts contemporary genetic structure in city rats. Peer Community in Ecology, 100044. https://doi.org/10.24072/pci.ecology.100044
Recommendation
Urban areas are expanding worldwide, and have become a dominant part of the landscape for many species. Urbanization can fragment pre-existing populations of vulnerable species leading to population declines and the loss of connectivity. On the other hand, expansion of urban areas can also facilitate the spread of human commensals including pests. Knowledge of the features of cityscapes that facilitate gene flow and maintain diversity of pests is thus key to their management and eradication.
Cities are complex mosaics of natural and manmade surfaces, and habitat quality is not only influenced by physical aspects of the cityscape but also by socioeconomic factors and human behaviour. Constant development means that cities also change rapidly in time; contemporary urban life reflects only a snapshot of the environmental conditions faced by populations. It thus remains a challenge to identify the features that actually drive ecology and evolution of populations in cities [1]. While several studies have highlighted strong urban clines in genetic structure and adaption [2], few have considered the influence of factors beyond physical aspects of the cityscape or historical processes.
In this paper, Stragier et al. [3] sought to identify the current and past features of the cityscape and socioeconomic factors that shape genetic structure and diversity of the house mouse (Mus musculus domesticus) in Dakar, Senegal. The authors painstakingly digitized historical maps of Dakar from the time of European settlement in 1862 to present. The authors found that the main spatial genetic cline was best explained by historical cityscape features, with higher apparent gene flow and genetic diversity in areas that were connected earlier to initial European settlements. Beyond the main trend of spatial genetic structure, they found further evidence that current features of the cityscape were important. Specifically, areas with low vegetation and poor housing conditions were found to support large, genetically diverse populations. The authors demonstrate that their results are reproducible using several statistical approaches, including modeling that explicitly accounts for spatial autocorrelation.
The work of Stragier et al. [3] thus highlights that populations of city-dwelling species are the product of both past and present cityscapes. Going forward, urban evolutionary ecologists should consider that despite the potential for rapid evolution in urban landscapes, the signal of a species’ colonization can remain for generations.
References
[1] Rivkin, L. R., Santangelo, J. S., Alberti, M. et al. (2019). A roadmap for urban evolutionary ecology. Evolutionary Applications, 12(3), 384-398. doi: 10.1111/eva.12734
[2] Miles, L. S., Rivkin, L. R., Johnson, M. T., Munshi‐South, J. and Verrelli, B. C. (2019). Gene flow and genetic drift in urban environments. Molecular ecology, 28(18), 4138-4151. doi: 10.1111/mec.15221
[3] Stragier, C., Piry, S., Loiseau, A., Kane, M., Sow, A., Niang, Y., Diallo, M., Ndiaye, A., Gauthier, P., Borderon, M., Granjon, L., Brouat, C. and Berthier, K. (2020). Interplay between historical and current features of the cityscape in shaping the genetic structure of the house mouse (Mus musculus domesticus) in Dakar (Senegal, West Africa). bioRxiv, 557066, ver. 4 peer-reviewed and recommended by PCI Ecology. doi: 10.1101/557066
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
no declaration
Evaluation round #2
DOI or URL of the preprint: https://doi.org/10.1101/557066
Version of the preprint: 2 (October 3, 2019)
Author's Reply, 29 Jan 2020
Decision by Michelle DiLeo, posted 05 Dec 2019
Dear Authors,
First, I would like to sincerely apologise for the delay in handling your manuscript. The revision came in while I was out of office, and finding a third reviewer to evaluate the methodological additions to the manuscript took longer than expected.
I have now received three reviews on the revised manuscript. I applaud the authors on the thoroughness of this revision. It is clear that a tremendous amount of work went into this, and I believe that the new analyses show that the results are relatively robust, and also introduce approaches that I have not seen used elsewhere in landscape-genetic studies. The reviewers for the most part share my enthusiasm, however, I agree with reviewer 3 that the extensive additions have now led to a decrease in readability. I unfortunately I cannot recommend the manuscript at this time, however I believe only minor changes are needed before the manuscript can be accepted. The suggested changes do not require new analyses, and can be addressed through additional clarification and by moving some of the new methods and results to the supplementary material.
I invite the authors to respond carefully to all reviewer comments. In particular, please work on increasing the readability. For example, both the RF methods produced similar results, so please only present one and move the methods and results of the other to the supplement. Similarly, the authors can consider omitting the model selection on the INLA results and instead just present the full model, with interpretation following from a comparison of coefficients.
Thank you and I look forward to seeing the revised manuscript.
Sincerely, Michelle DiLeo
Reviewed by anonymous reviewer 1, 19 Nov 2019
I have revised the new version of this work and I find that the authors have satisfactorily addressed my concerns, in particular adding the simulation study to assess the issue of sample size bring more robustness to the results found. I have only very few minor comments. Overall I think this is an interesting study, which contributes to understand the relationship between urbanization and population dynamics of invasive fauna.
Specific comments: Ln223: of a minimum of 20 captured… Ln270: groups Ln304: explain why these selection of radius (300, 600, 1000, 1500), does it have to do with is social organization, dispersal, etc. It is more interesting to know why this minimum and maximum value. Ln514: R2 to R2 (some are in capital and others no) In the conclusion, maybe it can be added a sentence of what can be done next to bring this type of studies to more practical recommendations for rodent control, o to other research avenues the authors consider important to address in the near future.
https://doi.org/10.24072/pci.ecology.100049.rev21Reviewed by , 25 Oct 2019
Stragier et al. have revised their previous manuscript to include a population-based analysis approach, which shows results consistent with previous results. In addition, the authors did a simulation to assess whether MAPI can be biased due to small number of sampling sites and they did resampling comparison to show that the original results are present in even smaller number of sampling sites. Furthermore, the authors have provided additional analysis on land cover variables and their relationship to genetic estimates. All in all, this brings quite a lot new results to the manuscript, but also makes the case much stronger.
My main concern was about the relationship between historical versus contemporary aspects of urban environment and I am happy with the additional analysis, explanations and revisions that the authors did.
I think the discussion needs still a bit polishing (some suggestions below): the flow is sub-optimal and some paragraphs are not very clear as they contain a lot of different ideas. I would suggest using less names of variables and more emphasis on the conclusions. For example, “Result from the RF and CAR models also suggested that the land cover class “Spontaneous” had a negative impact on genetic differentiation” does not open a new paragraph in very accessible way. State first your claim and they argue for it.
Some minor comments: - In abstract, maybe outline shortly what it actually means that “current genetic structure reflects the interplay between the historical dynamics of urbanization and the variation of contemporary urban habitats” – what are your actual results, i.e., something similar to line 693-695. - Lines 581-594: this paragraph reads much more technical than previous paragraphs – less about individual variables and more about broader implications might bring it more into line with other parts of discussion - Line 595: Maybe start this paragraph with what you are trying to say, now the beginning reads as a result. Also, this paragraph is too long for all the contents you are trying to put there. - Line 666: I would not describe your sample size small, but rather what you say in parenthesis - suboptimal sampling sites for spatial coverage.
https://doi.org/10.24072/pci.ecology.100049.rev22Reviewed by Torsti Schulz, 28 Nov 2019
Evaluation round #1
DOI or URL of the preprint: https://doi.org/10.1101/557066
Author's Reply, 03 Oct 2019
Please find in "reply" PDF file the answers to comments. The MS have been so deeply modified that so no track-change would be useful. Many thanks!
Decision by Michelle DiLeo, posted 02 Apr 2019
Dear Authors,
I have now received two reviews on your manuscript. In general, both reviewers were positive about the potential of this manuscript to make an interesting contribution, and I share their enthusiasm. That said, the reviewers had concerns and suggest several areas for improvement, which I would like to see addressed before I consider recommendation. While I would like to see replies to all of the reviewers concerns, below I highlight the areas that I think need the most work.
In particular, I share the concern that the site-level sampling strategy is perhaps not ideal, and that the implications of this should be discussed. It seems that MAPI is more suited to individual-based sampling (or at least finer-scale sampling) and I am interested to know if the results are sensitive to the resolution of the generated MAPI surface. How do the results of MAPI compare to analyses done with only the fourteen sampled sites?
Second, I echo the reviewer comment about the ability to tease apart the effects of historical versus contemporary aspects of the cityscape. It seems that these variables would be correlated, and it should be made abundantly clear to the reader how this problem was considered. I suspect that if there were no problems with collinearity, this might be easily addressed by reporting pairwise correlations and variance inflation factors for all variables.
Third, I agree that the discussion could use some work and that too much space is reserved for discussing the colonization history, which in my opinion is not the main or most interesting message of the paper.
Finally, I would be interested to see if genetic diversity correlates with the same aspects of the cityscape compared to genetic differentiation. Genetic diversity and differentiation can be driven by different processes and I think both are important when considering the practical applications of this work. I understand that with 14 sites you are limited to what can be included in a single model, but does genetic diversity show any strong univariate relationships with cityscape features beyond just the European settlement vs ancient village dichotomy?
Thank you for this interesting submission and I look forward to seeing your revision.
Sincerely, Michelle DiLeo
Reviewed by anonymous reviewer 1, 01 Apr 2019
The objective of this work was to investigate the influence of historical and contemporary stages of urbanization on genetic structure and differentiation of the house mouse (Mus musculus domesticus) in Dakar, Senegal. For this the authors employ data on D-loop sequences and 15 microsatellite markers for 14 sampling sites representing different stages of urbanization and habitats and applied spatial genetic analyses using a recent genetic spatial network method, MAPI, and conditional autoregressive models. Overall I found this manuscript interesting given the management implications of pest control of the results, which is a good example of basic science and a link with practical applications. The manuscript in general is well written, the objectives were clearly stated, and methods are sound. However my major concern is regarding the spatial distribution of sampling, which was focused in few sampling points giving a clustered sampling for a species that rather is likely found all over the city. I would like that the authors explain/address how their choice of sampling may bias results of an analysis that is based on building an spatial planar network that estimates spatial variations in pairwise genetic differentiation. The authors mentioned the challenges of sampling in this type of studies but did not mention the potential bias it can bring for landscape genetic analyses. I have other specific comments regarding clarification of methods and results, and presentation of figures.
See below:
Abstract: - Archetypal commensals: consider using other term, I don’t think this is clear for everyone - Remove in mice - I think to be more precise instead of analyzing the influence of historical and current features on genetic structure, it`s on genetic differentiation as the analyses requires genetic distances which are a proxy of gene flow. I don’t find the last sentences of the abstract regarding methods and results very useful, please be more specific on what type of analyses were carried and mention the most important results.
Introduction: - In the first it is mentioned that urbanization leads to genetic isolation, but there are already evidence showing that species can adapt and be benefited for urbanization processes, thus I think this paragraph needs a better overall framework, specially since the focus is on a species that has somehow benefited and demographically expanded due to urbanization.
Methods:
- The last paragraph of the section regarding classification of urban maps and cover classes can be shortened by omitting the details of all existing cover classes, which as explained later not all were explained. It would be more useful to know why the six-classes of urban habitats based on socioeconomic profiles were used, that is, which are the biological relevance for the dispersal or occurrence of the species? Also, I don’t have much clear if all vegetation types were considered just as one class, and if it is, does it mean that the species does not have any preference for a specific type of vegetation?
- Why the number of 20 samples as target? Also, it is not specified if adults or juveniles were collected and if the species is easily distinguished from other rodent fauna in the city.
- Specify the base pairs amplified for the D-loop
-Mitochondrial analysis: It is not clear with how many sequences the haplotype network was constructed. In the introduction it is mentioned that the D-loop data was also used to investigate the geographic origin of the house mouse with data from its entire distribution, so I don’t have clear if the haplotype network was the only analyses performed to address this objective. I think performing phylogenetic trees are also needed for answering this.
-Microsatellite analyses: (1) For what specific purpose the kinship coefficient was estimated? For performing landscape genetic analyses is important to eliminate related samples (sibs, half sibs), which I think can be better evaluated with other software, such as CERVUS. (2) Accuracy of effective population size estimates depends highly on sample size, does n=20 is enough to obtain accurate estimates given than this species is quite abundant where it occurs? (3) Sampling was not spatially uniform, but rather clustered in few sampling points within locations with different urban history. How does sampling bias or in this case, having very few points within a spatial grid where the species can occur everywhere may affect the results? This is because the hexagonal grid resulting from MAPI shows a fine mapping of genetic discontinuities (gradient colors), but this the spacing between some sampling locations is not small.
Results: -The first sentence can be omitted. -Improve flow of the sentence about the number of D-loop haplotypes. -The information of haplotype frequency by city regions is hard to follow as it is mainly descriptive, I think a figure will help, figure S3 should be in the main text. -What does it mean to have a kinship value f 0.2 in terms of relatedness? I still think a more specific relatedness analysis is needed, to know if related samples were taken and if need to be omitted for further analyses. -Please mention the allelic richness values for the two groups: ancient villages and the European settlement, just mentioning they are significant different, does not tell much about the relative difference. And, for the other diversity measures? The trend was the same, higher in the first European settlements? -Ne values are not mentioned in results - The removal or not of IDG was not mentioned in methods for the IBD test. Which is the explanation for performing this?
Discussion -I think results, discussion and the figure 2 will be easier to follow and understand if instead of naming urban 1 to urban 6, the name of what this classes represent is mentioned. For e.g, until discussion I knew that urban 5 refers to industrial areas.
Conclusion. -In my opinion the conclusion should state only stress the main finding and the implication of the results. The statement about the challenges and limitation of sampling in this type of studies should go into discussion and also adding what type of implications may have on the type of analyses used and the results found.
https://doi.org/10.24072/pci.ecology.100049.rev11Reviewed by , 26 Mar 2019
The authors describe a study where they examined variation in house mice mitochondrial and nuclear microsatellite markers in sites within the cityscape of Dakar, Senegal, with a differential urbanization history. They found patterns which reflect both urbanization history and current urban structure.
I find this manuscript very interesting and well-composed piece of urban ecology and population genetics. While rodents are one of the most notorious inhabitants of the urban landscape, from the point of view of material damage and pathogen transmission, there have been surprisingly little studies on their population ecology or genetics. Furthermore, while house mice is an old companion of humans, it has been less studied in urban environments than brown or black rats. Dakar seems to be a good case study of mouse population structure as it has both quite recent invasion of house mouse, and the city has also been going through a steady urban expansion. Thus this work might have not only important messages to the study of urban ecology and evolution, but also to more applied study of rodents in cityscape.
I find the manuscript convincing. I am not especially well-versed in population genetics, so I cannot evaluate their methods in depth, but I have no reason to suspect this work is not reliable. In general, this works relies on and nicely expands well-executed previous work on both house mouse genetics and urban history.
I have two major issues with the manuscript.
Firstly, it is unclear to me how the authors can differentiate between the effects of current urban structures and the historical structure of the urban landscape. I am not familiar with Dakar, but one would suspect that areas built at different time periods have different structural properties for house mouse and thus there would be correlation between time of urbanization and/or connection to urban area and the structural property of the area from the point of view of gene flow. Looking at the different patterns of prevalence of land covers, they do seem to partly correlate with the pattern of urban extension. It seems to me that the authors just assume that any genetic structure would be due to historic urban expansion. Why is that? Furthermore, the intriguing discrepancy between time since the connection to the early settlement and first occurrence of built-up areas suggests that there is more complex interplay of different aspects of urban structure than it first it seems.
Second issue is the structure and clarity of discussion: the authors now discuss in length the introduction of house mouse to Dakar, which seems secondary to what they have actually studied. It might make sense to just state that the genetic pattern is in line with the idea that mouse was introduced to first continental settlement at the south of Cap-Vert. In some parts of the discussion, the authors could make a clearer connection with their work and what they are arguing. For example, in the paragraph on trustworthiness the authors clearly have something important to say, specifically with their second point, but it is not clear how this is related the methods or results of this work. I would reorganize the contents of discussion and try to be more concise.
Minor issues: - In first paragraph of Introduction, the authors oppose “numerous species” and “commensal species”. Arguably also some commensal species can be spatially isolated in cityscape. Thus I find introducing commensal species as those which can easily disperse in urban landscape a bit strange. Maybe just state that some species can disperse more easily than others in urban landscape? - In second paragraph, yes, rodent control is costly (how costly?), but “weighs heavily on city’s budget” sounds like an overkill. My hunch is that more often than not, rodent control is just seen as essential “this has to be paid” part of budgets. - The first argument on the importance of spatiotemporal variation in gene flow seems to relate more actual movements of rodents than gene flow. If we know that rodents move from building to building, doing population genetics does not really give any additional help. The second and third argument are more on point. - In “Spatiotemporal pattern of urbanization”: what is ‘strict vegetation’? - In “Mitochondrial sequence analysis” the authors outline how they consider urban areas linked. I do not disagree with their approach, but it would be good to argue how their approach suits for house mouse by outlining what do we know (or do not know!) about house mouse biology/dispersal in urban environment. - The authors refer numerous times to applied aspects of their work, but it is not clear how straightforward it is. Is it sensible to have pretty much the whole downtown of Dakar as a simple target for eradication program, as I understood they are saying in the final paragraph of Discussion? - In conclusion, you refer to challenges outlined in Parsons et al. (2017) and cite those as the reasons for low sample size. Which of these challenges did you encounter? It might make sense to shortly outline those in Methods? Furthermore you suggest that collaboration can be the way forward – maybe put this in the discussion as it is not really your conclusion.
https://doi.org/10.24072/pci.ecology.100049.rev12









