
Hidden diversity: how genetic richness affects species diversity and ecosystem processes in freshwater ponds
Intraspecific diversity loss in a predator species alters prey community structure and ecosystem functions
Abstract
Recommendation: posted 05 November 2020, validated 16 November 2020
De Laender, F. (2020) Hidden diversity: how genetic richness affects species diversity and ecosystem processes in freshwater ponds. Peer Community in Ecology, 100064. 10.24072/pci.ecology.100064
Recommendation
Biodiversity loss can have important consequences for ecosystem functions, as exemplified by a large body of literature spanning at least three decades [1–3]. While connections between species diversity and ecosystem functions are now well-defined and understood, the importance of diversity within species is more elusive. Despite a surge in theoretical work on how intraspecific diversity can affect coexistence in simple community types [4,5], not much is known about how intraspecific diversity drives ecosystem processes in more complex community types. One particular challenge is that intraspecific diversity can be expressed as observable variation of functional traits, or instead subsist as genetic variation of which the consequences for ecosystem processes are harder to grasp.
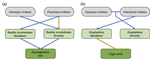
Raffard et al. [6] examined how intraspecific biodiversity loss in a consumer fish changes species diversity at lower trophic levels and ecosystem processes in pond mesocosms. An interesting feature of this experiment is that it crosses functional and genetic intraspecific diversity. To do so, Raffard and colleagues measured and genotyped European minnow (P. phoxinus) individuals sampled from streams across southern France. Combining these collected specimens into experimental ponds allowed them to control functional (population variance of body size) and genetic intraspecific richness (number of genotypes).
Effects on minnow biomass production were mostly small; biomass was significantly reduced only when lowering both functional and genetic richness. However, the consequences for lower trophic levels (zooplankton and macroinvertebrates) were more pronounced and – importantly – not intuitive. For instance, the macroinvertebrate community was less species-diverse at higher minnow functional richness. If minnows with different body sizes would be the main regulator factors [7] explaining macroinvertebrate interactions, one would expect a more diverse set of minnow body sizes (i.e. higher functional minnow richness) to permit higher instead of lower macroinvertebrate richness. At the same time, the macroinvertebrate community was more species-diverse at higher minnow genotype richness, which could indicate unobserved minnow traits determining macroinvertebrate diversity more than the usual suspects (functional consumer richness). Such unobserved traits could be behavioral traits, allowing for resource partitioning among fish.
The consequences of functional minnow diversity loss on zooplankton diversity were negative, as expected in case body size differences among fish would facilitate coexistence of their zooplankton prey, as explained above. However, this was only the case when genetic diversity was high, suggesting nonstraightforward interactive effects of observed and non-observed traits on prey diversity.
The effects of functional and genetic minnow diversity loss on invertebrate (macroinvertebrates and zooplankton) abundance were more consistent than for invertebrate diversity. This suggests again nonstraightforward relationships in this experimental ecosystem, but now between invertebrate diversity and abundance. When using abundance as a proxy for an ecosystem process (which the authors did not), this result illustrates that biodiversity loss in multitrophic communities can have consequences that are challenging to interpret, let alone predict [8,9]. Path analyses showed how the observed changes of invertebrate diversity and abundance co-determined decomposition, a key ecosystem function. These path analyses had highest explanatory power show when including both kinds of intraspecific diversity.
Taken together, the results by Raffard and colleagues suggest that genetic consumer richness can drive species diversity of connected trophic levels and ecosystem processes with similar magnitude as functional diversity. Indeed, the effects of genetic consumer richness were shown to be so strong as to compensate or exacerbate the loss of observed functional richness. The exact mechanisms explaining these effects remain to be identified, however. The possibility that fish grazing by fish with different (observed or not observed) traits regulates coexistence among invertebrate prey, for instance, would depend on how strong fish consumption feeds back on prey growth during a 30-week experiment. As the authors indicate, detailed studies on resource partitioning among consumers (e.g. using stable isotope labelling) can shed light on these matters. Doing so may address a more fundamental question, which is if the mechanisms linking intraspecific diversity to function are different from those linking interspecific diversity to function, and at what time scales.
References
[1] Tilman D, Downing JA (1994) Biodiversity and stability in grasslands. Nature, 367, 363–365. https://doi.org/10.1038/367363a0
[2] Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzig AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava DS, Naeem S (2012) Biodiversity loss and its impact on humanity. Nature, 486, 59–67. https://doi.org/10.1038/nature11148
[3] De Laender F, Rohr JR, Ashauer R, Baird DJ, Berger U, Eisenhauer N, Grimm V, Hommen U, Maltby L, Meliàn CJ, Pomati F, Roessink I, Radchuk V, Brink PJV den (2016) Reintroducing Environmental Change Drivers in Biodiversity–Ecosystem Functioning Research. Trends in Ecology & Evolution, 31, 905–915. https://doi.org/10.1016/j.tree.2016.09.007
[4] Hart SP, Schreiber SJ, Levine JM (2016) How variation between individuals affects species coexistence. Ecology Letters, 19, 825–838. https://doi.org/10.1111/ele.12618
[5] Barabás G, D’Andrea R (2016) The effect of intraspecific variation and heritability on community pattern and robustness. Ecology Letters, 19, 977–986. https://doi.org/10.1111/ele.12636
[6] Raffard A, Cucherousset J, Montoya JM, Richard M, Acoca-Pidolle S, Poésy C, Garreau A, Santoul F, Blanchet S (2020) Intraspecific diversity loss in a predator species alters prey community structure and ecosystem functions. bioRxiv, 2020.06.10.144337, ver. 3 peer-reviewed and recommended by PCI Ecology. https://doi.org/10.1101/2020.06.10.144337
[7] Pásztor L, Botta-Dukát Z, Magyar G, Czárán T, Meszéna G. Theory-Based Ecology: A Darwinian approach. Oxford University Press. https://doi.org/10.1093/acprof:oso/9780199577859.001.0001
[8] Binzer A, Guill C, Rall BC, Brose U (2016) Interactive effects of warming, eutrophication and size structure: impacts on biodiversity and food-web structure. Global Change Biology, 22, 220–227. https://doi.org/10.1111/gcb.13086
[9] Schwarz B, Barnes AD, Thakur MP, Brose U, Ciobanu M, Reich PB, Rich RL, Rosenbaum B, Stefanski A, Eisenhauer N (2017) Warming alters energetic structure and function but not resilience of soil food webs. Nature Climate Change, 7, 895–900. https://doi.org/10.1038/s41558-017-0002-z
The recommender in charge of the evaluation of the article and the reviewers declared that they have no conflict of interest (as defined in the code of conduct of PCI) with the authors or with the content of the article. The authors declared that they comply with the PCI rule of having no financial conflicts of interest in relation to the content of the article.
no declaration
Reviewed by Andrew Barnes, 03 Nov 2020
On a second reading, I am reminded that this is a very impressive, novel, and elegant experimental study that highlights some very exciting patterns around the influence of genetic diversity on ecosystem functioning in multitrophic systems! I am impressed with the careful revision by the authors and feel they have done a fantastic job of addressing almost all points. The only remaining issue I see is around the effect size display in the path models.
It is certainly a challenge to present results for paths from categorical variables, such as genotypic and functional richness in this study. However, I think that weighting based on p-values is highly problematic and misleading, as path width typically infers something about effect size (not statistical significance), for which p-values should be avoided. One possibility could be to use something like Cohen's f2, as this would give the partial effect where you have multiple predictors. I haven’t used this myself, so this might require some more investigation. However, an alternative would be to simply avoid weighting the paths altogether (which would be far preferable to weighting by p-value), and simply refer to reported statistics in table 1. In combination with Figure S5, this should give enough information about the relative importance of the different variables, and would give a much more honest presentation of effects in the path models. The only other addition I can think of that could support interpretation would be r-squared values (or pseudo r-squared for mixed effects models) for each response variable, to indicate how well the predictor sets explain variance in each response. I hope this is of some help and look forward to seeing the revised paper!
Evaluation round #1
DOI or URL of the preprint: 10.1101/2020.06.10.144337
Version of the preprint: 1
Author's Reply, 09 Oct 2020
Decision by Frederik De Laender, posted 12 Aug 2020
Both reviewers acknowledge that the contribution is novel and potentially of interest to a broad range of ecologists. One reviewer suggests adding reflections on other traits than body size. The other reviewer is more critical and lists a number of methodological concerns.
Additional requirements of the managing board:
As indicated in the 'How does it work?’ section and in the code of conduct, please make sure that:
-Data are available to readers, either in the text or through an open data repository such as Zenodo (free), Dryad or some other institutional repository. Data must be reusable, thus metadata or accompanying text must carefully describe the data.
-Details on quantitative analyses (e.g., data treatment and statistical scripts in R, bioinformatic pipeline scripts, etc.) and details concerning simulations (scripts, codes) are available to readers in the text, as appendices, or through an open data repository, such as Zenodo, Dryad or some other institutional repository. The scripts or codes must be carefully described so that they can be reused.
-Details on experimental procedures are available to readers in the text or as appendices.
-Authors have no financial conflict of interest relating to the article. The article must contain a "Conflict of interest disclosure" paragraph before the reference section containing this sentence: "The authors of this preprint declare that they have no financial conflict of interest with the content of this article." If appropriate, this disclosure may be completed by a sentence indicating that some of the authors are PCI recommenders: “XXX is one of the PCI XXX recommenders.”
Reviewed by Andrew Barnes, 24 Jul 2020
Reviewed by Jes Hines, 15 Jul 2020
A few decades of experiments have shown that, all other things being equal, communities with lower species diversity often have reduced ecosystem functioning and stability. Therefore, conserving and restoring species diversity is often touted as a key priority for science policy programs. However, the implications of these “biodiversity-ecosystem function” (BEF) experiments have been controversial because species do not go extinct randomly, and also because ecological stressors placed on populations may have strong consequences for ecosystem function that precede species extinction. For example, threatened species often have small or declining populations with low population genetic diversity and high inbreeding. Yet, there has been far less investigation into the ecosystem consequences of these declines.
Raffard et al present an elegant experiment where they test the consequences of changes in genotypic and functional diversity of a fish, Phoxinus phoxinus, on biodiversity and ecosystem functioning of fish and adjacent trophic levels. They make two important contribution to biodiversity-ecosystem functioning (BEF) literature. First, the authors seek to understand whether genetic diversity is a better proxy for differences in phenotypic expression of complementary but potentially unmeasured functional traits (e.g. body size), or whether measures of functional diversity are sufficient to capture effects of consumer variation on ecosystem functioning. Second, they use fish as their model organism. In a literature dominated by experiments reporting manipulation of plants, it is refreshing to see the ecosystem effects of a changes in larger consumer species also being documented.
They found that fish diversity consistently influenced ecosystem functioning by altering benthic invertebrate diversity, and also by altering zooplankton diversity but only when functional diversity was high. The experiment is carefully conceived and the treatments are well justified using several combinations of fish from wild populations with previously determined genotypic differences. The observed effects are likely robust as the experimental mesocosms were stocked with zooplankton and algae and natural colonization of invertebrates was also allowed. The natural colonization added an extra component of realism and stochasticity that is often missing from highly controlled experiments.
My only minor concern about the manuscript is that cryptic phenotypic diversity is emphasized as a mechanism underlying genotypic diversity. While I don’t doubt this is true, it seems that there should be several more functional traits that could be easily measured in addition to body size, which is the trait predominately emphasized in the text. It would be helpful to give some concrete examples of other traits, even if they are not explicitly measured in this experiment. Otherwise, discussion of the term cryptic diversity may seem unnecessarily vague.
The following comments are made only to show that my lack of major criticisms is not due to lack of reading the manuscript. The rationale for the experiment is well defined and well-justified within the scope of broader literature. The methods are informative. The statistical analyses are appropriate and clearly described. All tables and figures are useful and carefully prepared. The discussion thoughtfully evaluates the results. Relevant literature is cited discerningly.
Minor comments Line 335 typo: extend not extent Line 337 typo sustain(s)










