
NATER Chloé
- Department for Terrestrial Biodiversity, Norwegian Institute for Nature Research, Trondheim, Norway
- Biodiversity, Conservation biology, Demography, Freshwater ecology, Life history, Marine ecology, Population ecology, Spatial ecology, Metacommunities & Metapopulations, Statistical ecology, Terrestrial ecology
Recommendations: 0
Review: 1
Review: 1
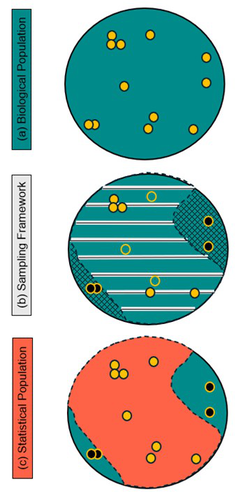
Using multiple datasets to account for misalignment between statistical and biological populations for abundance estimation
Diving into detection process to solve sampling and abundance issues in a cryptic species
Recommended by Guillaume Souchay based on reviews by Michael Schaub, Chloé Nater and 1 anonymous reviewerEstimating population parameters is critical for analysis and management of wildlife populations. Drawing inference at the population level requires a robust sampling scheme and information about the representativeness of the studied population (Williams et al. 2002). In their textbook, Williams et al. (see chapter 5, 2002) listed several sampling issues, including both temporal and spatial heterogeneity and especially imperfect detection. Several methods, either sampling-based or model-based can be used to circumvent these issues.
In their paper, Kissling et al. (2024) addressed the case of the Kittlitz’s murrelet (Brachyramphus brevirostris), a cryptic ice-associated seabird, combining spatial variation in its distribution, temporal variation in breeding propensity, imperfect detection and logistical challenges to access the breeding area. The Kittlitz’s murrelet is thus the perfect species to illustrate common issues and logistical difficulties to implement a standard sampling scheme.
The authors proposed a modelling framework unifying several datasets from different surveys to extract information on each step of the detection process: the spatial match between the targeted population and the sampled population, the probability of presence in the sample area, the probability of availability given presence in the sample area and finally, the probability of detection given presence and availability. All these components were part of the framework to estimate abundance and trend for this species.
They took advantage of a radiotracking survey during several years to inform spatial match and probability of presence. They performed a behavioural experiment to assess the probability of availability of murrelets given it was present in sampling area, and they used a conventional distance-sampling boat survey to estimate detection of individuals. This is worth noting that the most variable components were the probability of presence in the sample area, with a global mean of 0.50, and the probability of detection given presence and availability ranging from 0.49 to 0.77. The estimated trend for Kittlitz’s murrelet was negative and all the information gathered in this study will be useful for future conservation plan.
Coupling a decomposition of the detection process with different data sources was the key to solve problems raised by such “difficult” species, and the paper of Kissling et al. (2024) is a good way to follow for other species, allowing to inform the detection components for the targeted species - and also for our global understanding of detection process, and to infer about the temporal trend of species of conservation concern.
References
Williams, B. K., Nichols, J. D., and Conroy, M. J. (2002). Analysis and management of animal populations. Academic Press.
Michelle L. Kissling, Paul M. Lukacs, Kelly Nesvacil, Scott M. Gende, Grey W. Pendleton (2024) Using multiple datasets to account for misalignment between statistical and biological populations for abundance estimation. EcoEvoRxiv, ver.3 peer-reviewed and recommended by PCI Ecology https://doi.org/10.32942/X2W03T