
ARNAUD-HAOND Sophie
- MARBEC, Ifremer, SETE, France
- Biodiversity, Biogeography, Biological invasions, Conservation biology, Dispersal & Migration, Evolutionary ecology, Marine ecology, Molecular ecology
- recommender
Recommendations: 2
Review: 1
Recommendations: 2

Metabarcoding faecal samples to investigate spatiotemporal variation in the diet of the endangered Westland petrel (Procellaria westlandica)
The promise and limits of DNA based approach to infer diet flexibility in endangered top predators
Recommended by Sophie Arnaud-Haond based on reviews by Francis John Burdon and Babett GüntherThere is growing evidence of worldwide decline of populations of top predators, including marine ones (Heithaus et al, 2008, Mc Cauley et al., 2015), with cascading effects expected at the ecosystem level, due to global change and human activities, including habitat loss or fragmentation, the collapse or the range shifts of their preys. On a global scale, seabirds are among the most threatened group of birds, about one-third of them being considered as threatened or endangered (Votier& Sherley, 2017). The large consequences of the decrease of the populations of preys they feed on (Cury et al, 2011) points diet flexibility as one important element to understand for effective management (McInnes et al, 2017). Nevertheless, morphological inventory of preys requires intrusive protocols, and the differential digestion rate of distinct taxa may lead to a large bias in morphological-based diet assessments. The use of DNA metabarcoding on feces (or diet DNA, dDNA) now allows non-invasive approaches facilitating the recollection of samples and the detection of multiple preys independently of their digestion rates (Deagle et al., 2019). Although no gold standard exists yet to avoid bias associated with metabarcoding (primer bias, gaps in reference databases, inability to differentiate primary from secondary predation…), the use of these recent techniques has already improved the knowledge of the foraging behaviour and diet of many animals (Ando et al., 2020).
Both promise and shortcomings of this approach are illustrated in the article “Metabarcoding faecal samples to investigate spatiotemporal variation in the diet of the endangered Westland petrel (Procellaria westlandica)” by Quereteja et al. (2021). In this work, the authors assessed the nature and spatio-temporal flexibility of the foraging behaviour and consequent diet of the endangered petrel Procellaria westlandica from New-Zealand through metabarcoding of faeces samples.
The results of this dDNA, non-invasive approach, identify some expected and also unexpected prey items, some of which require further investigation likely due to large gaps in the reference databases. They also reveal the temporal (before and after hatching) and spatial (across colonies only 1.5km apart) flexibility of the foraging behaviour, additionally suggesting a possible influence of fisheries activities in the surroundings of the colonies. This study thus both underlines the power of the non-invasive metabarcoding approach on faeces, and the important results such analysis can deliver for conservation, pointing a potential for diet flexibility that may be essential for the resilience of this iconic yet endangered species.
References
Ando H, Mukai H, Komura T, Dewi T, Ando M, Isagi Y (2020) Methodological trends and perspectives of animal dietary studies by noninvasive fecal DNA metabarcoding. Environmental DNA, 2, 391–406. https://doi.org/10.1002/edn3.117
Cury PM, Boyd IL, Bonhommeau S, Anker-Nilssen T, Crawford RJM, Furness RW, Mills JA, Murphy EJ, Österblom H, Paleczny M, Piatt JF, Roux J-P, Shannon L, Sydeman WJ (2011) Global Seabird Response to Forage Fish Depletion—One-Third for the Birds. Science, 334, 1703–1706. https://doi.org/10.1126/science.1212928
Deagle BE, Thomas AC, McInnes JC, Clarke LJ, Vesterinen EJ, Clare EL, Kartzinel TR, Eveson JP (2019) Counting with DNA in metabarcoding studies: How should we convert sequence reads to dietary data? Molecular Ecology, 28, 391–406. https://doi.org/10.1111/mec.14734
Heithaus MR, Frid A, Wirsing AJ, Worm B (2008) Predicting ecological consequences of marine top predator declines. Trends in Ecology & Evolution, 23, 202–210. https://doi.org/10.1016/j.tree.2008.01.003
McCauley DJ, Pinsky ML, Palumbi SR, Estes JA, Joyce FH, Warner RR (2015) Marine defaunation: Animal loss in the global ocean. Science, 347, 1255641. https://doi.org/10.1126/science.1255641
McInnes JC, Jarman SN, Lea M-A, Raymond B, Deagle BE, Phillips RA, Catry P, Stanworth A, Weimerskirch H, Kusch A, Gras M, Cherel Y, Maschette D, Alderman R (2017) DNA Metabarcoding as a Marine Conservation and Management Tool: A Circumpolar Examination of Fishery Discards in the Diet of Threatened Albatrosses. Frontiers in Marine Science, 4, 277. https://doi.org/10.3389/fmars.2017.00277
Querejeta M, Lefort M-C, Bretagnolle V, Boyer S (2021) Metabarcoding faecal samples to investigate spatiotemporal variation in the diet of the endangered Westland petrel (Procellaria westlandica). bioRxiv, 2020.10.30.360289, ver. 4 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2020.10.30.360289
Votier SC, Sherley RB (2017) Seabirds. Current Biology, 27, R448–R450. https://doi.org/10.1016/j.cub.2017.01.042
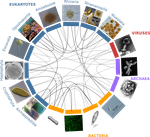
A community perspective on the concept of marine holobionts: current status, challenges, and future directions
Marine holobiont in the high throughput sequencing era
Recommended by Sophie Arnaud-Haond and Corinne Vacher based on reviews by Sophie Arnaud-Haond and Aurélie TasiemskiThe concept of holobiont dates back to more than thirty years, it was primarily coined to hypothesize the importance of symbiotic associations to generate significant evolutionary novelties. Quickly adopted to describe the now well-studied system formed by zooxanthella associated corals, this concept expanded much further after the emergence of High-Throughput Sequencing and associated progresses in metabarcoding and metagenomics.
Holobionts – defined as the association between an individual host and its microbiota - are now increasingly described at sea and on land. The opinion article by Dittami et al. [1] provides a synthetic overview of marine holobionts. It retraces the history of the holobiont concept, recalls the main mechanisms underlying the association between hosts and microbial communities, highlights the influence of these symbioses on marine ecosystem functioning, and outlines current tools and future lines of research.
In particular, the article discusses some particularities of marine systems, such as the strong connectivity allowing an exchange of microorganisms and chemical signals between and within holobionts.
The authors advocate the need to bridge the gap between large scale exploration studies and smaller scale mechanistic studies, by conducting interdisciplinary research (combining physiology, biochemistry, ecology, experimentation and computational modeling) on some keystone holobionts.
Finally, one strength of the paper by Dittami et al. [1] is that it places the concept of the holobiont in an applied research framework. Several possible applications of knowledge on host-microbiota interactions are suggested, both in the field of aquaculture and that of monitoring the health of marine ecosystems. This article contains all the necessary elements for someone who would like to jump into the study of the holobionths in the marine world.
References
[1] Dittami SM, Arboleda E, Auguet J, Bigalke A, Briand E, Cardenas P, Cardini U, Decelle J, Engelen AH, Eveillard D, Gachon CMM, Griffiths SM, Harder T, Kayal E, Kazamia E, Lallier FH, Medina M, Marzinelli E, Morganti T, Núñez Pons L, Prado S, Pintado J, Saha M, Selosse M, Skillings D, Stock W, Sunagawa S, Toulza E, Vorobev A, Leblanc C, Not F. (2020). A community perspective on the concept of marine holobionts: current status, challenges, and future directions. Zenodo, ver. 4 peer-reviewed and recommended by PCI Ecology. doi: 10.5281/zenodo.3696771
Review: 1

A community perspective on the concept of marine holobionts: current status, challenges, and future directions
Marine holobiont in the high throughput sequencing era
Recommended by Sophie Arnaud-Haond and Corinne Vacher based on reviews by Sophie Arnaud-Haond and Aurélie TasiemskiThe concept of holobiont dates back to more than thirty years, it was primarily coined to hypothesize the importance of symbiotic associations to generate significant evolutionary novelties. Quickly adopted to describe the now well-studied system formed by zooxanthella associated corals, this concept expanded much further after the emergence of High-Throughput Sequencing and associated progresses in metabarcoding and metagenomics.
Holobionts – defined as the association between an individual host and its microbiota - are now increasingly described at sea and on land. The opinion article by Dittami et al. [1] provides a synthetic overview of marine holobionts. It retraces the history of the holobiont concept, recalls the main mechanisms underlying the association between hosts and microbial communities, highlights the influence of these symbioses on marine ecosystem functioning, and outlines current tools and future lines of research.
In particular, the article discusses some particularities of marine systems, such as the strong connectivity allowing an exchange of microorganisms and chemical signals between and within holobionts.
The authors advocate the need to bridge the gap between large scale exploration studies and smaller scale mechanistic studies, by conducting interdisciplinary research (combining physiology, biochemistry, ecology, experimentation and computational modeling) on some keystone holobionts.
Finally, one strength of the paper by Dittami et al. [1] is that it places the concept of the holobiont in an applied research framework. Several possible applications of knowledge on host-microbiota interactions are suggested, both in the field of aquaculture and that of monitoring the health of marine ecosystems. This article contains all the necessary elements for someone who would like to jump into the study of the holobionths in the marine world.
References
[1] Dittami SM, Arboleda E, Auguet J, Bigalke A, Briand E, Cardenas P, Cardini U, Decelle J, Engelen AH, Eveillard D, Gachon CMM, Griffiths SM, Harder T, Kayal E, Kazamia E, Lallier FH, Medina M, Marzinelli E, Morganti T, Núñez Pons L, Prado S, Pintado J, Saha M, Selosse M, Skillings D, Stock W, Sunagawa S, Toulza E, Vorobev A, Leblanc C, Not F. (2020). A community perspective on the concept of marine holobionts: current status, challenges, and future directions. Zenodo, ver. 4 peer-reviewed and recommended by PCI Ecology. doi: 10.5281/zenodo.3696771