
DE WIT Rutger
Recommendations: 0
Review: 1
Review: 1
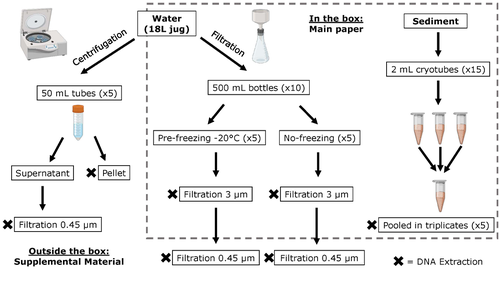
Different approaches to processing environmental DNA samples in turbid waters have distinct effects for fish, bacterial and archaea communities.
Processing environmental DNA samples in turbid waters from coastal lagoons
Recommended by Claudia Piccini based on reviews by David Murray-Stoker and Rutger De WitCoastal lagoons are among the most productive natural ecosystems on Earth. These relatively closed basins are important habitats and nursery for endemic and endangered species and are extremely vulnerable to nutrient input from the surrounding catchment; therefore, they are highly susceptible to anthropogenic influence, pollution and invasion (Pérez-Ruzafa et al., 2019). In general, coastal lagoons exhibit great spatial and temporal variability in their physicochemical water characteristics due to the sporadic mixing of freshwater with marine influx. One of the alternatives for monitoring communities or target species in aquatic ecosystems is the environmental DNA (eDNA), since overcomes some limitations from traditional methods and enables the investigation of multiple species from a single sample (Thomsen and Willerslev, 2015). In coastal lagoons, where the water turbidity is highly variable, there is a major challenge for monitoring the eDNA because filtering turbid water to obtain the eDNA is problematic (filters get rapidly clogged, there is organic and inorganic matter accumulation, etc.).
The study by Turba et al. (2023) analyzes different ways of dealing with eDNA sampling and processing in turbid waters and sediments of coastal lagoons, and offers guidelines to obtain unbiased results from the subsequent sequencing using 12S (fish) and 16S (Bacteria and Archaea) universal primers. They analyzed the effect on taxa detection of: i) freezing or not prior to filtering; ii) freezing prior to centrifugation to obtain a sample pellet; and iii) using frozen sediment samples as a proxy of what happens in the water. The authors propose these different alternatives (freeze, do not freeze, sediment sampling) because they consider that they are the easiest to carry out. They found that freezing before filtering using a 3 µm pore size filter had no effects on community composition for either primer, and therefore it is a worthwhile approach for comparison of fish, bacteria and archaea in this kind of system. However, significantly different bacterial community composition was found for sediment compared to water samples. Also, in sediment samples the replicates showed to be more heterogeneous, so the authors suggest increasing the number of replicates when using sediment samples. Something that could be a concern with the study is that the rarefaction curves based on sequencing effort for each protocol did not saturate in any case, this being especially evident in sediment samples. The authors were aware of this, used the slopes obtained from each curve as a measure of comparison between samples and considering that the sequencing depth did not meet their expectations, they managed to achieve their goal and to determine which protocol is the most promising for eDNA monitoring in coastal lagoons. Although there are details that could be adjusted in relation to this item, I consider that the approach is promising for this type of turbid system.
References
Pérez-Ruzafa A, Campillo S, Fernández-Palacios JM, García-Lacunza A, García-Oliva M, Ibañez H, Navarro-Martínez PC, Pérez-Marcos M, Pérez-Ruzafa IM, Quispe-Becerra JI, Sala-Mirete A, Sánchez O, Marcos C (2019) Long-Term Dynamic in Nutrients, Chlorophyll a, and Water Quality Parameters in a Coastal Lagoon During a Process of Eutrophication for Decades, a Sudden Break and a Relatively Rapid Recovery. Frontiers in Marine Science, 6. https://doi.org/10.3389/fmars.2019.00026
Thomsen PF, Willerslev E (2015) Environmental DNA – An emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation, 183, 4–18. https://doi.org/10.1016/j.biocon.2014.11.019
Turba R, Thai GH, Jacobs DK (2023) Different approaches to processing environmental DNA samples in turbid waters have distinct effects for fish, bacterial and archaea communities. bioRxiv, 2022.06.17.495388, ver. 2 peer-reviewed and recommended by Peer Community in Ecology. https://doi.org/10.1101/2022.06.17.495388