

Endophytic fungi are expected to be hyperdiverse in tropical forests, and here is an article exploring their diversity, hidden in Rubiaceae leaves, in two old-growth forests of Costa Rica. Humberto Castillo-González et al. not only described their diversity, but also test for the impact of leaf development stage, tissue origin, and site location. They distinguish the different fungal lineages and do identify distinct indicators, showing that specialization of endophytic fungi could be related to other factors in tropical forests.
This article is a great example of fungal ecology in the tropics, interacting at fine and large scale with a diversity of hosts. It also invites to discuss the high specialization observed in the tropics, and the ecology of old-growth forests in Costa Rica.
References
Humberto Castillo-González, Jason C. Slot, Stephanie Yarwood, Priscila Chaverri (2025) Exploring Rubiaceae fungal endophytes across contrasting tropical forests, tree tissues, and developmental stages. bioRxiv, ver.3 peer-reviewed and recommended by PCI Ecology https://doi.org/10.1101/2024.02.13.580172
DOI or URL of the preprint: https://doi.org/10.1101/2024.02.13.580172
Version of the preprint: 2
Dear Mélanie:
Thank you very much for the positive feedback.
In the revised version, we have modified the sentence in the Introduction, added some keywords, and added a reference for the methods.
Please let me know if anything else needs to be fixed.
Best regards,
Priscila
Dear colleagues,
our last reviewer did a great revision, and I do agree all point were adressed. A last sentence might be corrected in the introduction, and perhaps clarifying if you developped your protocols for sampling or not (not mandatory). All other points are really improved, I'll prepare my recommender text while you finish the edition of your preprint.
Thanks for your patience, best,
Mélanie
 , 17 Dec 2024
, 17 Dec 2024DOI or URL of the preprint: https://doi.org/10.1101/2024.02.13.580172
Version of the preprint: 1
Dear colleagues,
First of all, I apologize for the delay in handling your revision, experts of tropical endophytes are rare ! Both reviewers did a great job, adding positive comments to your already well shaped manuscript.
I noticed in both case, reviewers suggested to precise hypothesis, methodology, and analysis to validate the importance of spatial patterns and beta diversity changes. Those are really detailled and minor changes that could improve your manuscript, I hope you would appreciate those comments. Both reviewers discussed the importance of defining individuals vs host effects, and the limit of defining specialization vs generalism. interesting references are mentionned, as well as perspectives on core microbiome analysis (focusing on what's stable, rather than variations !).
Your study represents an important piece of work on the topic, and the tropics, as endophytes diversity remains little explored at such a scale and in old-growth forests. Do not hesitate to resubmit with these recommendations !
 , 06 Jun 2024
, 06 Jun 2024Overall comment:
This study presents the fungal endophyte diversity and composition within the Rubiaceae family. Fungal endophytes communities are compared across host species, between contrasting locations, between different tissue types, and development stages. This manuscript is very interesting, and the findings shall contribute to generate interest among the readers, especially considering that knowledge on fungal endophytes in tropical rain forests is still scarce. Having said that, I think that there is space for improving the manuscript and I offer my comments/recommendations.
Evaluation of the different components of the article:
Title :
The article title could be enhanced regarding the hypotheses explored: "Exploring Rubiaceae fungal endophytes across contrasting tropical forests, tree tissues and development stages."
Abstract
The abstract presents the main goal of the study and the associated results. I have a few minor remarks:
L29-30: This sentence could be deleted.
L30: I suggest to detail more the method as such: "Sapwood, mature and young leaves were collected from 47 Rubiaceae species in old-growth forests in Golfito and Guanacaste, Costa Rica. Fungal diversity and composition was assessed using metabarcoding of the ITS2 nrDNA region."
Introduction
The introduction explains the motivation for the study, eventhough an accent could be made on why the authors chose to study fungal endophytes. Endophytes are broader than just fungi, maybe could add details to this point. Hypotheses need also to be refined to enhance clarity.
L50-51: It is worth mentioning here that there are also studies on root fungal endophytes, not only concerning mycorrhizal and free-living forms: "but
traditionally understudied compared to root fungal endophtyes, mycorrhizal and free-living forms".
L53-54: While, the introduction on the role of endophytes is smoothly progressing, the mention here feels somewhat abrupt and it needs to be more developped.
L56: More details are needed on horizontal transfer and why it is considered the main colonization process (for the authors) since vertical transfer is extremely important as well.
L72: There is a jump from Rubiaceae family to a specific genera, and more details are needed to explain why the focus on Palicourea?
L77-85: Emphasize that you are studying fungal endophytes. Endophytes are much broader than only fungi. Why fungal endophytes can be more clearly enhance through out the introduction.
L78: The first hypothesis is clear and interesting. What is the expect outcome, ex higher diversity in Golfito compared to Guanacaste due to higher rainfalls in this region?
L80: The second hypothesis could be more refined, and more details on the expectation can be given. It can be broken down into more than one. First on plant and trees, second on tissue type and development stage, for example.
L82-83 : No hypotheses were given concerning fungal host specialization and this could be enhanced. To what question or hypotheses do the CLAM test and indicator taxa respond to?
L84: What is meant by "processes"? If it is diversity and composition, these are not processes. Community processes refer to selection, dispersion, drift, speciation. Perhaps a more precise word is needed here.
Materials and methods
This section can be more detailed and here are my specific comments:
L88 Study site: Geographical coordinates can be added to the text and also to the Table1. It would also be helpful to provide a brief description of the Figure 1 in the text.
L100: To be more precise, talk about invididuals instead of plants
L100:The selection suggests a broad sampling approach, how were the 47 species selected? What was the criteria?
L101: Here "plants" means "species" no? Start the sentence such as "The taxonomy was confirmed by... "
L103: Were the branched sampled in the upper or lower crown of the tree?
L104-105: For the differences in development stages, why would three nodes be sufficient to separate new and mature leaves?
L106: Again, individuals ?
L110: Was this protocol for leaf surface sterilization developped specifically for this study or is it a standard protocol?
L113: Less details are provided concerning the sapwood procedure. I'm curious to know why the sapwood sampled were singed? (slightly burned?) I have never worked with sapwood sampled before.
L116: Even if the experimental procedures were outsourced or used in kits for the DNA extraction, it would be helpful to give the key steps for each procedure: DNA extraction, the PCR, what were the negative and positive controls, which reverse and forward primers were used. Why were primers flTS7 and ITS4 suitable ?
L131-133: What cutoffs were used during the curation process to filter quality, trim and remove low abundance reads? Provide secifics about the quality thresholds applied.
L141: Which version of R was used?
L142: Give more details to why alpha diversity indices were transformed to Hill effective species numbers. Perhaps cite that this was shown to provide more robust estimates of diversity (Jost et al., 2010)
L147: Following a PERMANOVA, was the homogeneity of dipersion checked to ensure that differences in PERMANOVA were not due to differences in disperion? It can easily be checked using the betadisper function in the vegan package.
L148-151: The sentence is dense, perhaps break it down. Restate also what were the 6 categories. Chose either categories or habitat.
Results
Consider structuring the results with sub-section headings to enhance the cohesion and flow, maybe according to the hypotheses? Otherwise, yes the results were described and interpreted correctly.
L162 the first line of the results section could easily be moved at the end of the materials & method, at the end of the paragraph ASV classification and taxa assigment.
L178-179: English needs rephrasing such as: "Regarding the development stage, the taxo richness (q=0) was significantly higher in mature leaves compared to new leaves.
L188: It is hear mentioned that dispersion homogeneity was check but not mentioned in the materials and methods.
L198: Perhaps the paragraph on the taxonomy could be merged after L167, at the end of the first paragraph.
Tables and figures
The tables and figures are overall very nicely done.
Figure 1: L792: Is it necessary to add "Made per request.." Perhaps add it to the materials and methods text or in the acknowledgments. There is no legend concerning the colors of the climatic subregions, eventhough some are described in the legend text.
Figure 2: L799: More details in the legend could be given: "... (A) different locations, Golfito and Guanacaste, (B) different tissues, mature and new leaves and sapwood, ..."
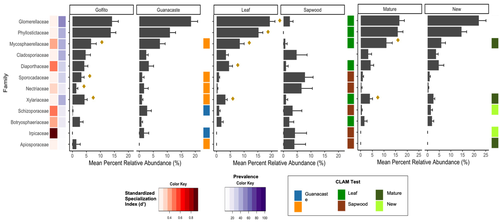
Figure 3: L806: Great figure, very clear! Just a suggestion to make it even more clearer: I found the CLAM test legend confusing. I would only say : “habitat specialization is indicated by the CLAM test”. Because no colors means habitat generalization, no? Also, make the legend key wider for all legend text to spell out correctly.
Table 1: Geographical coordinates can be added to the text and also to the Table1.
Discussion
I overall would suggest the structure of the discussion to be devided with sub-section titles to help the flow of the discussion. I suggest to first summarize the main findings and then address clearly each hypothesis in the order it was presented in the introduction. I also did not see how the first part of the second hypothesis (concerning the difference between plants and trees) was assessed and discussed, but perhaps I missed something.
L247: I don't fully agree with this statement since species identity could drive the community, eventhough the results in this study did not find fungal endophyte communities to cluster according to taxonomy. It does not mean that plant taxonomy does not shape the community, the interpretation here is too abrupt.
L254: I don't really understand the link between the results concerning H1 and the temporal distribution of endophytes.
L260: "higher organisms" do you mean macro-organisms? Maybe a more precise word is needed here.
L262-64: How are the environemental conditions related to higher variability in fungal endophyte communities. A clearer link is needed here to understand the interpretation, not just citing the Table 1 and Figure 1.
L266-270: Perhaps other unmeasured environmental factors could explain the distribution ? (ex. Humidity, wind, soil characteristics, forest structure and composition…)
L268 Do you mean variation of composition ?
L269: neutral processes are not just dispersion limitation, but also consider ecological drift and diversification (Vellend et al., 2010; Nemergut et al., 2013)
L270: I do not completely agree, there is also vertical transmission. The plant does not germinate without an initial microbiota. The future horizontal colonizing endophytes have also to compete with already present microorganisms.
L274-75: I suggest to keep all the idea about distance decay in one unique paragraph.
L282-288: The paragraph is overall confusing with the first sentence saying that both locations were dominated by host generalists, but the second part mentions a comparison with higher specialists. The mention "host generalist" refers to the trees or fungi? It is confusing. Revision of english wording is needed too. I suggest to develop the idea of host generalist, their potential influence/interaction with microorganisms, compared to host specialists if this is the idea of the paragraph. Or develop the biology, lifestyle, physiology of fungi generalists vs specialists.
L298: What do the two different compartment leaf vs spawood offer in terms of niche requirements for the endophytes to thrive?
L342-345: Small contradiction here: It was mentioned that leaves are considered oligotrophic environments (L289-290), and here is written high nutrient availability.
Discussion: do they talk about roots, other microbes? Endophtyes are not only fungi, maybe broader with bacteria, protists etc.. and talk about roots? An importance source of horizontal transfer is from the soil to the roots to the leaves.
L381: I would summarize the main results as done in this paragraph, at the very beginning of the discussion. And perhaps end the discussion opening to the challenges in the conservation /agriculture .
L346-47: This is the expectation worth mentioning in the introduction for the second hypothesis.
L346-52: I would simplify this sentence and nuance the meaning behing since this study did not evaluate changes in genetic traits, tissue chemistry, topology, plant hormonal, physiological properties, physical characteristics to see the direct impact on the community composition.
L360: It is really true that plants in the tropics do not necessarily experience strong selective pressures to develop sophisticated chemical defenses? It may be worth checking this assumption again. It's not my area of expertise so I'm probably learning something new.
L361-63: This is actually a very interesting results that could maybe be related to the notion of a core microbiome of the Rubiaceae family: a stable fungal community within the host across habitats, taxonomy species, and development stages. Maybe an idea to explore.
L381: I would not include leaves and sapwood as part of the phyllosphere, I would dissociate them.
References
L411 and L533: Check the reference format.
The authors have produced a manuscript which details an important and understudied component of tropical ecosystems. By collecting a significant number of specimens across two broad geographic regions, at different leaf ages and tissue types they are able to show the importance of these regions for endophyte diversity, and begin to show how this diversity is structured. It is well written and contains clear support for points made with the use of well put together figures in addition to supplementary methods. Although I think this study is worthy of publication, it could be significantly improved by adding in a little more detail with regards to how they reach their hypotheses, a few areas of methodology lacking clarity and an acknowledgment of where this study is limited. These points are detailed further below.
Title and abstract are clear
Introduction
57 -would benefit from more info on environmental and geographical factors influencing this. You mention these in your hypotheses but it would be good to highlight studies where distance or topography have been revealed as potentially important in structuring endophyte communities. For example (Suryanarayanan, Murali, & Venkatesan, 2002; Suryanarayanan, Venkatesan, & Murali, 2003; Zimmerman & Vitousek, 2012; Cordier et al., 2012; Izuno, Kanzaki, Artchawakom, Wachrinrat, & Isagi, 2016). Many of these feature in your discussion but they could be clarified here to guide the reader to how you reach your hypotheses
72-79 - your hypotheses line up with the literature but appear somewhat out of nowhere, can you refer to other studies where authors have looked at diversity patterns of endophytes within different plant species e.g Arnold et al. 2000; Donald et al. 2020 or different tissues Carroll 1988
Methods
100-101 - very limited replication within species, I.e. only one tree per species. In addition, the combination of species is not equivalent across sites. This means you cannot test for a species effect on diversity as all you would show here is differences between individual trees. In addition, any inference of differences in location could be down to the different combination of host tree species but your analysis cannot account for this. Your results are still of interest for such an understudied region, but this limitation could result in an over inference of importance. This needs acknowledging in the discussion, but also reappraising in your introduction and hypotheses. In your analysis and results, you cluster at the family level where you would have more replication.
105 - can you clarify for people like me not good at maths, how many leaves per tree?
113- more detail required on “singed” method
123- how did you have any prc or extraction replication from your individual leaf samples?
131-134 - you mention numerous filtering steps but there is no discussion of what thresholds these were set at, or a record of how many reads / asv s were removed at each step. This numbers should be stated since they give an indication as to the quality of your dataset. Did you have any blanks / extraction or pcr controls? Aside from sequence errors such as chimeras, how does this method account for potential contaminants?
156- can you clarify what you mean by variable classes? This would help the reader understand what you mean when you refer to a specialist or indicator
Results
176- how confident can you be in differences in endophytes from different tissues when you use different methods to conduct the extractions. The tissues themselves may be trickier to extract rna from?
209-210 do you have enough replicates of host tree species to determine if they are specialists? See Novotny et al., 2002
264- I would caution against over inference of the role of environmental controls here given you only have two blocks of habitat sampled rather than any specific reported range of experienced environment with sufficient replication
Discussion
You need to have a section which acknowledges the limitations of this study. Although you reveal lots of great patterns relating to differences across two regions in CR, there should be more caution or caveats made when referring to detected environmental effects or what your analysis o specialist / generalist taxa really can show.
More modifications to your methods to address the comments above will require the modification of your discussion to account for this. Otherwise, points are well supported with reference to a broad pool of relevant literature demonstrating how 5e research fits into the current state of art well.